Meru co-ordinates spindle orientation with cell polarity and cell cycle progression
- PMID: 40169811
- PMCID: PMC12084343
- DOI: 10.1038/s44318-025-00420-5
Meru co-ordinates spindle orientation with cell polarity and cell cycle progression
Abstract
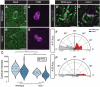
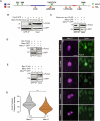
Correct mitotic spindle alignment is essential for tissue architecture and plays an important role in cell fate specification through asymmetric cell division. Spindle tethering factors such as Drosophila Mud (NuMA in mammals) are recruited to the cell cortex and capture astral microtubules, pulling the spindle in the correct orientation. However, how spindle tethering complexes read the cell polarity axis and how spindle attachment is coupled to mitotic progression remains poorly understood. We explore these questions in Drosophila sensory organ precursors (SOPs), which divide asymmetrically to give rise to epidermal mechanosensory bristles. We show that the scaffold protein Meru, which is enriched at the posterior cortex by the Frizzled/Dishevelled planar cell polarity complex, in turn recruits Mud, linking the spindle tethering and polarity machineries. Furthermore, Cyclin A/Cdk1 associates with Meru at the posterior cortex, promoting the formation of the Mud/Meru/Dsh complex via Meru and Dsh phosphorylation. Thus, Meru couples spindle orientation with cell polarity and provides a cell cycle-dependent cue for spindle tethering.
Keywords: Asymmetric Cell Division; Cell Polarity; Development; Drosophila; Spindle Orientation.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures












Similar articles
-
The Fz-Dsh planar cell polarity pathway induces oriented cell division via Mud/NuMA in Drosophila and zebrafish.Dev Cell. 2010 Nov 16;19(5):740-52. doi: 10.1016/j.devcel.2010.10.004. Dev Cell. 2010. PMID: 21074723 Free PMC article.
-
Cortical Cyclin A controls spindle orientation during asymmetric cell divisions in Drosophila.Nat Commun. 2022 May 17;13(1):2723. doi: 10.1038/s41467-022-30182-1. Nat Commun. 2022. PMID: 35581185 Free PMC article.
-
Inscuteable regulates the Pins-Mud spindle orientation pathway.PLoS One. 2012;7(1):e29611. doi: 10.1371/journal.pone.0029611. Epub 2012 Jan 10. PLoS One. 2012. PMID: 22253744 Free PMC article.
-
Epithelial polarity and spindle orientation: intersecting pathways.Philos Trans R Soc Lond B Biol Sci. 2013 Sep 23;368(1629):20130291. doi: 10.1098/rstb.2013.0291. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24062590 Free PMC article. Review.
-
Molecular pathways regulating mitotic spindle orientation in animal cells.Development. 2013 May;140(9):1843-56. doi: 10.1242/dev.087627. Development. 2013. PMID: 23571210 Free PMC article. Review.
References
-
- Albertson R, Chabu C, Sheehan A, Doe CQ (2004) Scribble protein domain mapping reveals a multistep localisation mechanism and domains necessary for establishing cortical polarity. J Cell Sci 117(25):6061–6070 - PubMed
-
- Albertson R, Doe CQ (2003) Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol 5(2):166–170 - PubMed
-
- Bastock R, Strutt H, Strutt D (2003) Strabismus is asymmetrically localised and binds to Prickle and Dishevelled during Drosophila planar polarity patterning. Development 130:3007–3014 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

