Integrative analysis of a novel signature incorporating metabolism and stemness-related genes for risk stratification and assessing clinical outcomes and therapeutic responses in lung adenocarcinoma
- PMID: 40170009
- PMCID: PMC11963273
- DOI: 10.1186/s12885-025-13984-6
Integrative analysis of a novel signature incorporating metabolism and stemness-related genes for risk stratification and assessing clinical outcomes and therapeutic responses in lung adenocarcinoma
Abstract
Background: Metabolism and stemness-related genes (msRGs) are critical in the development and progression of lung adenocarcinoma (LUAD). Nevertheless, reliable prognostic risk signatures derived from msRGs have yet to be established.
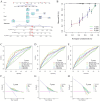
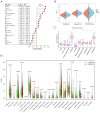
Methods: In this study, we downloaded and analyzed RNA-sequencing and clinical data from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases. We employed univariate and multivariate Cox regression analyses, along with least absolute shrinkage and selection operator (LASSO) regression analysis, to identify msRGs that are linked to the prognosis of LUAD and to develop the prognostic risk signature. The prognostic value was evaluated using Kaplan-Meier analysis and log-rank tests. We generated receiver operating characteristic (ROC) curves to evaluate the predictive capability of the prognostic signature. To estimate the relative proportions of infiltrating immune cells, we utilized the CIBERSORT algorithm and the MCPCOUNTER method. The prediction of the half-maximal inhibitory concentration (IC50) for commonly used chemotherapy drugs was conducted through ridge regression employing the "pRRophetic" R package. The validation of our analytical findings was performed through both in vivo and in vitro studies.
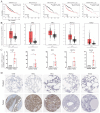
Results: A novel five-gene prognostic risk signature consisting of S100P, GPX2, PRC1, ARNTL2, and RGS20 was developed based on the msRGs. A risk score derived from this gene signature was utilized to stratify LUAD patients into high- and low-risk groups, with the former exhibiting significantly poorer overall survival (OS). A nomogram was constructed incorporating the risk score and other clinical characteristics, showcasing strong capabilities in estimating the OS rates for LUAD patients. Furthermore, we observed notable differences in the infiltration of various immune cell subtypes, as well as in responses to immunotherapy and chemotherapy, between the low-risk and high-risk groups. Results from gene set enrichment analysis (GSEA) and in vitro studies indicated that the prognostic signature gene ARNTL2 influenced the prognosis of LUAD patients, primarily through the activation of the PI3K/AKT/mTOR signaling pathway.
Conclusions: Utilizing this gene signature for risk stratification could help with clinical treatment management and improve the prognosis of LUAD patients.
Keywords: Bioinformatics; Immune infiltration; Lung adenocarcinoma; Metabolism and stemness-related genes signature.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Our research adheres to the Declaration of Helsinki. The studies involving human participants were reviewed and approved by the Ethics Committee of Medical School of Henan University, China (HUSOM-2018-282). Informed consent was obtained from all subjects involved in the study. The animal study was reviewed and approved by the Ethic Committee of Medical School of Henan University. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Integration of single-cell and bulk RNA sequencing to identify a distinct tumor stem cells and construct a novel prognostic signature for evaluating prognosis and immunotherapy in LUAD.J Transl Med. 2025 Feb 22;23(1):222. doi: 10.1186/s12967-025-06243-6. J Transl Med. 2025. PMID: 39987127 Free PMC article.
-
Stratification of Lung Adenocarcinoma Patients Based on In Silico and Immunohistochemistry Analyses of Oxidative Stress-Related Genes.Cancer Biother Radiopharm. 2025 Jan;40(1):11-21. doi: 10.1089/cbr.2024.0094. Epub 2024 Jul 1. Cancer Biother Radiopharm. 2025. PMID: 38949986
-
Multi-omics analysis-based clinical and functional significance of a novel prognostic and immunotherapeutic gene signature derived from amino acid metabolism pathways in lung adenocarcinoma.Front Immunol. 2024 Dec 13;15:1361992. doi: 10.3389/fimmu.2024.1361992. eCollection 2024. Front Immunol. 2024. PMID: 39735553 Free PMC article.
-
Development and validation of a robust immune-related prognostic signature in early-stage lung adenocarcinoma.J Transl Med. 2020 Oct 7;18(1):380. doi: 10.1186/s12967-020-02545-z. J Transl Med. 2020. PMID: 33028329 Free PMC article.
-
Fatty acid metabolism prognostic signature predicts tumor immune microenvironment and immunotherapy, and identifies tumorigenic role of MOGAT2 in lung adenocarcinoma.Front Immunol. 2024 Oct 16;15:1456719. doi: 10.3389/fimmu.2024.1456719. eCollection 2024. Front Immunol. 2024. PMID: 39478862 Free PMC article.
References
-
- Lovly CM. Expanding horizons for treatment of Early-Stage lung cancer. N Engl J Med. 2022;386:2050–1. - PubMed
-
- Das A, Rewari S, Kanaujia BK, Deswal SS, Gupta RS. Analytical investigation of a triple surrounding gate germanium source metal–oxide–semiconductor field-effect transistor with step graded channel for biosensing applications. Int J Numer Model Electron Networks Devices Fields 2023; 36.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

