Accelerating high-concentration monoclonal antibody development with large-scale viscosity data and ensemble deep learning
- PMID: 40170162
- PMCID: PMC12128653
- DOI: 10.1080/19420862.2025.2483944
Accelerating high-concentration monoclonal antibody development with large-scale viscosity data and ensemble deep learning
Abstract
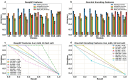
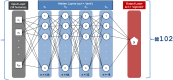
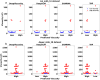
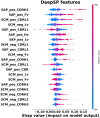
Highly concentrated antibody solutions are necessary for developing subcutaneous injections but often exhibit high viscosities, posing challenges in antibody-drug development, manufacturing, and administration. Previous computational models were only limited to a few dozen data points for training, a bottleneck for generalizability. In this study, we measured the viscosity of a panel of 229 monoclonal antibodies (mAbs) to develop predictive models for high concentration mAb screening. We developed DeepViscosity, consisting of 102 ensemble artificial neural network models to classify low-viscosity (≤20 cP) and high-viscosity (>20 cP) mAbs at 150 mg/mL, using 30 features from a sequence-based DeepSP model. Two independent test sets, comprising 16 and 38 mAbs with known experimental viscosity, were used to assess DeepViscosity's generalizability. The model exhibited an accuracy of 87.5% and 89.5% on both test sets, respectively, surpassing other predictive methods. DeepViscosity will facilitate early-stage antibody development to select low-viscosity antibodies for improved manufacturability and formulation properties, critical for subcutaneous drug delivery. The webserver-based application can be freely accessed via https://devpred.onrender.com/DeepViscosity.
Keywords: Antibody viscosity; ensemble deep learning; high-concentration formulations; monoclonal antibodies.
Conflict of interest statement
AL, MH, CC, AD, GK, VS, MY, MP, MB, JC, AH, MK, MS, AG, and NM are all AstraZeneca employees and may or may not hold AstraZeneca stock. MD was an AstraZeneca employee and may or may not hold AstraZeneca stock and is currently at Xaira Therapeutics and may or may not hold Xaira Therapeutics stock.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
