ORF1ab codon frequency model predicts host-pathogen relationship in orthocoronavirinae
- PMID: 40170904
- PMCID: PMC11958986
- DOI: 10.3389/fbinf.2025.1562668
ORF1ab codon frequency model predicts host-pathogen relationship in orthocoronavirinae
Abstract
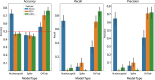
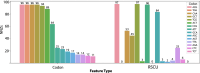
Predicting phenotypic properties of a virus directly from its sequence data is an attractive goal for viral epidemiology. Here, we focus narrowly on the Orthocoronavirinae clade and demonstrate models that are powerfully predictive for a human-pathogen phenotype with 76.74% average precision and 85.96% average recall on the withheld test set groups, using only Orf1ab codon frequencies. We show alternative examples for other viral coding sequences and feature representations that do not perform well and discuss what distinguishes the models that are performant. These models point to a small subset of features, specifically 5 codons, that are critical to the success of the models. We discuss and contextualize how this observation may fit within a larger model for the role of translation in virus-host agreement.
Keywords: bioinformactics; feature selection; genotype-to-phenotype; machine learning; viruses.
Copyright © 2025 Davis and Russell.
Conflict of interest statement
Authors PD and JR were employed by MRIGlobal.
Figures
Similar articles
-
Predicting viral host codon fitness and path shifting through tree-based learning on codon usage biases and genomic characteristics.Sci Rep. 2025 Apr 10;15(1):12251. doi: 10.1038/s41598-025-91469-z. Sci Rep. 2025. PMID: 40211017 Free PMC article.
-
The evolutionary and genetic patterns of African swine fever virus.Infect Genet Evol. 2024 Aug;122:105612. doi: 10.1016/j.meegid.2024.105612. Epub 2024 May 31. Infect Genet Evol. 2024. PMID: 38824981
-
Prediction of virus-host infectious association by supervised learning methods.BMC Bioinformatics. 2017 Mar 14;18(Suppl 3):60. doi: 10.1186/s12859-017-1473-7. BMC Bioinformatics. 2017. PMID: 28361670 Free PMC article.
-
Attenuation of Human Respiratory Viruses by Synonymous Genome Recoding.Front Immunol. 2019 Jun 4;10:1250. doi: 10.3389/fimmu.2019.01250. eCollection 2019. Front Immunol. 2019. PMID: 31231383 Free PMC article. Review.
-
Targeting Virus-host Protein Interactions: Feature Extraction and Machine Learning Approaches.Curr Drug Metab. 2019;20(3):177-184. doi: 10.2174/1389200219666180829121038. Curr Drug Metab. 2019. PMID: 30156155 Review.
References
LinkOut - more resources
Full Text Sources

