Integrative Multi-Omics and Routine Blood Analysis Using Deep Learning: Cost-Effective Early Prediction of Chronic Disease Risks
- PMID: 40171841
- PMCID: PMC12165040
- DOI: 10.1002/advs.202412775
Integrative Multi-Omics and Routine Blood Analysis Using Deep Learning: Cost-Effective Early Prediction of Chronic Disease Risks
Abstract
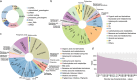
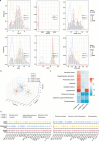
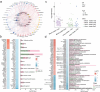
Chronic noncommunicable diseases (NCDS) are often characterized by gradual onset and slow progression, but the difficulty in early prediction remains a substantial health challenge worldwide. This study aims to explore the interconnectedness of disease occurrence through multi-omics studies and validate it in large-scale electronic health records. In response, the research examined multi-omics data from 160 sub-healthy individuals at high altitude and then a deep learning model called Omicsformer is developed for detailed analysis and classification of routine blood samples. Omicsformer adeptly identified potential risks for nine diseases including cancer, cardiovascular conditions, and psychiatric conditions. Analysis of risk trajectories from 20 years of large clinical patients confirmed the validity of the group in preclinical risk assessment, revealing trends in increased disease risk at the time of onset. Additionally, a straightforward NCDs risk prediction system is developed, utilizing basic blood test results. This work highlights the role of multiomics analysis in the prediction of chronic disease risk, and the development and validation of predictive models based on blood routine results can help advance personalized medicine and reduce the cost of disease screening in the community.
Keywords: multi‐omics; pathway discovery; potential risk classification.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
All authors declare no competing interests.
Figures



References
-
- C. O. WHO , Air Quality Guidelines for Europe 2020, 91.
-
- World Health Organization , World health statistics 2023: monitoring health for the SDGs, Sustainable Development Goals, World Health Organization, Geneva: 2023.
-
- Crosby D., Bhatia S., Brindle K. M., Coussens L. M., Dive C., Emberton M., Esener S., Fitzgerald R. C., Gambhir S. S., Kuhn P., Rebbeck T. R., Balasubramanian S., Science 2022, 375, eaay9040. - PubMed
-
- Hunter D. J., Reddy K. S., N. Engl. J. Med. 2013, 369, 1336. - PubMed
-
- Bhardwaj N., Wodajo B., Spano A., Neal S., Coustasse A., Health Care Manag. 2018, 37, 90. - PubMed
MeSH terms
Grants and funding
- Medical Engineering Laboratory of Chinese PLA General Hospital
- 2022ZD0209103/National Key R&D Program of China
- 2021ZD0140406/Ministry of Science and Technology of the People's Republic of China
- 62325604/National Natural Science Foundation of China
- 62276271/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
