Characterization of the Two-Domain Peptide Binding Mechanism of the Human CGRP Receptor for CGRP and the Ultrahigh Affinity ssCGRP Variant
- PMID: 40172014
- PMCID: PMC12004451
- DOI: 10.1021/acs.biochem.4c00812
Characterization of the Two-Domain Peptide Binding Mechanism of the Human CGRP Receptor for CGRP and the Ultrahigh Affinity ssCGRP Variant
Abstract
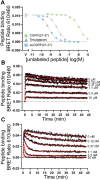
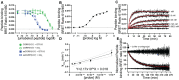
Calcitonin gene-related peptide (CGRP) is a 37-amino acid neuropeptide that functions in pain signaling and neuroimmune communication. The CGRP receptor, CGRPR, is a class B GPCR that is a drug target for migraine headache and other disorders. Here, we used nanoBRET receptor binding and cAMP biosensor signaling assays and theoretical modeling to characterize the CGRPR "two-domain" peptide binding mechanism. Single-site extracellular domain (ECD)-binding and two-site ECD/transmembrane domain (TMD)-binding peptides were examined for CGRP and a high-affinity variant "ssCGRP" with modifications in the C-terminal region. Wildtype and ssCGRP(27-37) bound the ECD with affinities of 1 μM and 0.5 nM, and residence times of 5 s and 8 min, respectively. The (8-37) antagonist fragments had affinities of 100 nM for wildtype and 0.5 nM for ss and exhibited behavior consistent with two-site ECD/TMD binding. ssCGRP(8-37) had a residence time of 76 min. CGRP(1-37) agonist had 25-fold higher affinity for the G protein-coupled state of the CGRPR (Ki = 3 nM) than the uncoupled state (Ki = 74 nM), and elicited short-duration cAMP signaling. In contrast, ssCGRP(1-37) had similar strong affinities for both receptor states (Ki = 0.2 to 0.25 nM), and induced long-duration signaling. An equilibrium reaction network mathematical model of CGRPR activation that includes peptide and G protein binding was developed. This captured wildtype CGRP binding experiments well, but the ssCGRP binding properties were not fully reproduced, suggesting that it may exhibit a distinct binding mechanism. Together, these results advance our quantitative understanding of the CGRPR two-domain mechanism and support the ss variants as potential long-acting therapeutics.
Copyright © 2025 The Authors. Published by American Chemical Society. This publication is licensed under CC-BY 4.0.
Conflict of interest statement
The authors declare the following competing financial interest(s): AP is inventor on a patent covering the ssCGRP variant. The other authors report no conflicts.
Figures






Similar articles
-
Virtual drug repurposing study for the CGRPR identifies pentagastrin and leuprorelin as putative candidates.J Mol Graph Model. 2022 Nov;116:108254. doi: 10.1016/j.jmgm.2022.108254. Epub 2022 Jun 18. J Mol Graph Model. 2022. PMID: 35803082
-
Relative Antagonism of Mutants of the CGRP Receptor Extracellular Loop 2 Domain (ECL2) Using a Truncated Competitive Antagonist (CGRP8-37): Evidence for the Dual Involvement of ECL2 in the Two-Domain Binding Model.Biochemistry. 2017 Aug 1;56(30):3877-3880. doi: 10.1021/acs.biochem.7b00077. Epub 2017 Jul 18. Biochemistry. 2017. PMID: 28691801
-
Selective CGRP and adrenomedullin peptide binding by tethered RAMP-calcitonin receptor-like receptor extracellular domain fusion proteins.Protein Sci. 2013 Dec;22(12):1775-85. doi: 10.1002/pro.2377. Epub 2013 Oct 19. Protein Sci. 2013. PMID: 24115156 Free PMC article.
-
Multiple affinity and guanine nucleotide sensitive forms of the calcitonin gene related peptide (CGRP) receptor.Can J Physiol Pharmacol. 1995 Jul;73(7):968-73. doi: 10.1139/y95-134. Can J Physiol Pharmacol. 1995. PMID: 8846438 Review.
-
Interaction of calcitonin-gene-related peptide with its receptors.Biochem Soc Trans. 2002 Aug;30(4):451-5. doi: 10.1042/bst0300451. Biochem Soc Trans. 2002. PMID: 12196113 Review.
Cited by
-
Determinants of Improved CGRP Peptide Binding Kinetics Revealed by Enhanced Molecular Simulations.bioRxiv [Preprint]. 2025 Jun 17:2025.06.13.659569. doi: 10.1101/2025.06.13.659569. bioRxiv. 2025. PMID: 40667178 Free PMC article. Preprint.
-
Copper only SOD repeat proteins likely act as an extracellular superoxide dismutase in oyster antioxidant defense.Sci Rep. 2025 Jul 1;15(1):20465. doi: 10.1038/s41598-025-06156-w. Sci Rep. 2025. PMID: 40594389 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

