Heat tolerance and genetic adaptations in Caenorhabditis briggsae: insights from comparative studies with Caenorhabditis elegans
- PMID: 40172097
- PMCID: PMC12135185
- DOI: 10.1093/genetics/iyaf061
Heat tolerance and genetic adaptations in Caenorhabditis briggsae: insights from comparative studies with Caenorhabditis elegans
Abstract
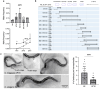
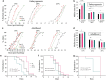
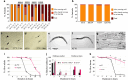
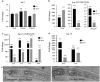
Temperature tolerance varies widely across species and plays a crucial role in shaping physiological and evolutionary adaptations. Here, we investigate thermal stress responses in Caenorhabditis briggsae and Caenorhabditis elegans using multiple isolates. Our results demonstrate that C. briggsae exhibits enhanced survival, growth, and reproduction at elevated temperatures compared to C. elegans. The increased heat resistance was evident from the L1 larval stage. Notably, C. briggsae isolates from both tropical and temperate regions were equally resistant to heat stress, suggesting that elevated thermal tolerance is an intrinsic feature of this species. To explore the molecular genetic basis of thermal tolerance, we examined expression of heat shock regulators. Transcriptional analysis revealed that C. briggsae mounts a rapid and robust heat shock response, with CBG19186, the closest ortholog of C. eleganshsp-16.2, showing higher induction and faster recovery dynamics. The peak expression of hsp-16.2/CBG19186 occurred at a temperature 2°C higher in C. briggsae than in C. elegans. These findings provide the first in vivo evidence of temperature differences in the transcriptional response of a single protein between the 2 species, suggesting that C. briggsae has evolved a higher thermal limit for key molecular processes, likely contributing to its ability to withstand extreme temperatures. Despite its superior thermal resistance, C. briggsae showed higher sensitivity to oxidative, osmotic, and endoplasmic reticulum stress, suggesting a potential fitness trade-off. Our findings demonstrate significant differences in stress sensitivities between the 2 nematodes, providing a foundation for further investigations into the molecular and evolutionary mechanisms underlying their stress responses.
Keywords: HSF-1; HSP-16.2; WormBase; heat shock protein; heat stress; nematode; thermal tolerance.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflicts of interest.
Figures
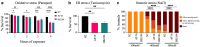

References
-
- Altun ZF, Hall DH. 2009. Introduction. In: Altun ZF, Herndon LA, Wolkow CA, Crocker C, Lints R, Hall DH, editors. WormAtlas. doi: 10.3908/wormatlas.1.1. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

