Vibrio cholerae O47 associated with a cholera-like diarrheal outbreak concurrent with seasonal cholera in Bangladesh
- PMID: 40172221
- PMCID: PMC12039230
- DOI: 10.1128/msphere.00831-24
Vibrio cholerae O47 associated with a cholera-like diarrheal outbreak concurrent with seasonal cholera in Bangladesh
Abstract
The Ganges delta of the Bay of Bengal is a recognized hotspot for the emergence and spread of novel variants of Vibrio cholerae. Despite being a diverse species, very little information is available concerning environmental and human-associated aspects of V. cholerae serogroups, other than the two major epidemic-related serogroups O1 and O139. This represents a crucial gap in understanding the spectrum of diversity, ecology, and epidemiology of the species influencing the dynamics of global cholera. In this study, we describe an emerging variant of V. cholerae displaying the antigenic property of serogroup O47, associated with a cholera-like outbreak in coastal Bangladesh where cholera has been endemic for centuries. This outbreak coincides with a rise in cases of cholera caused by V. cholerae O1, as well as frequency of isolation of serogroups O47 and O1 from the environment. The V. cholerae O47 isolates proved clonal in nature, and their genome biology revealed distinct features, with respect to multidrug resistance (MDR), serogroup-specific genes, genomic island combinations, and overall phylogenetic properties. Genome comparison confirmed the absence of canonical virulence factors of V. cholerae O1 and O139, namely, cholera toxin (CTX) and toxin-co-regulated pili (TCP), and the presence of putative virulence factors including type 3 secretion system (T3SS) and an MDR pseudo-compound transposon, carrying genes for macrolide resistance and extended spectrum beta-lactamase. Results of the study suggest that V. cholerae O47 could represent an emerging Vibrio pathogen with the potential to spread virulence and antimicrobial resistance traits impacting the management of cholera-like diseases.IMPORTANCEDespite the global insurgence of human diseases caused by Vibrios in recent years, most research focuses only on the O1 serogroup of V. cholerae, leaving a significant gap concerning the environmental and human-associated aspects of other serogroups found in nature. Although other serogroups are often found associated with sporadic diarrhea cases, in 1992-1993, a massive cholera-like diarrhea epidemic was initiated by a "non-O1" serogroup, namely, O139 that temporally displaced O1 from endemic cholera in the Bay of Bengal villages of Bangladesh and India, highlighting the potential threat they might pose. This study describes yet another emerging variant of V. cholerae, displaying the antigenic property of serogroup O47, associated with a cholera-like outbreak in a coastal locality in Bangladesh. Findings of the study offer critical insights into the genome biology of V. cholerae O47 and its potential implications for understanding their ecology and epidemiology of cholera-like diseases.
Keywords: NOVC; Vibrio cholerae; antimicrobial resistance; cholera; genome analysis; genomic island; outbreak; serogroup.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
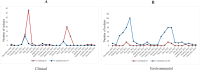


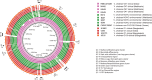

References
-
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Lee JH, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A, Nair GB, Colwell RR. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci USA 106:15442–15447. doi:10.1073/pnas.0907787106 - DOI - PMC - PubMed
-
- Murase K, Arakawa E, Izumiya H, Iguchi A, Takemura T, Kikuchi T, Nakagawa I, Thomson NR, Ohnishi M, Morita M. 2022. Genomic dissection of the Vibrio cholerae O-serogroup global reference strains: reassessing our view of diversity and plasticity between two chromosomes. Microb Genom 8:mgen000860. doi:10.1099/mgen.0.000860 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
