Type II grass carp reovirus utilizes autophagosomes for viroplasm formation and subclinical persistent infection
- PMID: 40172227
- PMCID: PMC12090803
- DOI: 10.1128/jvi.00352-25
Type II grass carp reovirus utilizes autophagosomes for viroplasm formation and subclinical persistent infection
Abstract
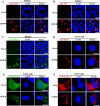
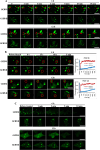
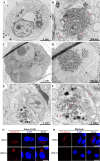
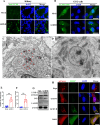
Grass carp reovirus (GCRV) is the most virulent pathogen within the genus Aquareovirus, belonging to the family Spinareoviridae. GCRV is categorized into three genotypes, with type II (GCRV-II) being the predominant strain circulating in China. Reoviruses are known to replicate and assemble in cytoplasmic inclusion bodies termed viroplasms; however, information regarding the formation of GCRV-II viroplasms and their specific roles in virus infection remains largely unknown. In this study, we investigated the formation and characteristics of viroplasms during GCRV-II infection. Immunofluorescence and confocal microscopy indicate that GCRV-II infection induces the formation of viroplasms, with the nonstructural protein NS79 being the key protein responsible for this process. Live-cell imaging and fluorescence recovery after photobleaching assays reveal that GCRV-II viroplasms lack liquid-like properties. Transmission electron microscopy confirms that GCRV-II viroplasms are membranous structures. Notably, we demonstrate that GCRV-II infection induces autophagy and the formation of autophagosomes and that GCRV-II utilizes these autophagosomes for viroplasm formation and virion assembly. Furthermore, we found that GCRV-II uses autophagosomes to evade the host immune system, establishing subclinical persistent infection. GCRV-II also employs autophagosomes for nonlytic release and viral spread. Collectively, these findings highlight distinctive characteristics of GCRV-II viroplasms compared to those of other animal reoviruses, offering valuable insights for the prevention and control of this virus.IMPORTANCEGrass carp reovirus (GCRV) is categorized into three genotypes, with GCRV-II being the most prevalent in China. Despite reoviruses being known for their replication and assembly in viroplasms, the specifics of GCRV-II viroplasm formation and its role in infection were unclear. Our study demonstrates that GCRV-II infection triggers the formation of viroplasms, primarily mediated by the nonstructural protein NS79. GCRV-II viroplasms are membranous structures that lack liquid-like properties, which are significantly different from the viroplasms of other reoviruses. Notably, our research unveils that GCRV-II infection induces autophagy and utilizes autophagosomes for viroplasm formation and virion assembly. Furthermore, we also confirm that GCRV-II utilizes autophagosomes for subclinical persistent infection, nonlytic release, and viral spread. Our results indicate that GCRV-II hijacks autophagosomes to form viroplasms and complete its life cycle. The characteristics of GCRV-II are significantly different from those of other animal reoviruses, providing important information for prevention and control of this virus.
Keywords: autophagosomes; autophagy; nonlytic release; nonstructural protein NS79; subclinical persistent infection; type II grass carp reovirus; viral spread; viroplasms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Liquid-liquid phase separation is essential for reovirus viroplasm formation and immune evasion.J Virol. 2024 Sep 17;98(9):e0102824. doi: 10.1128/jvi.01028-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194247 Free PMC article.
-
Grass Carp Reovirus Induces Formation of Lipid Droplets as Sites for Its Replication and Assembly.mBio. 2022 Dec 20;13(6):e0229722. doi: 10.1128/mbio.02297-22. Epub 2022 Nov 29. mBio. 2022. PMID: 36445081 Free PMC article.
-
ATG5 negatively regulates grass carp reovirus-induced immune-inflammatory response by degrading RIG-I and MDA5.J Virol. 2025 Jul 22;99(7):e0034425. doi: 10.1128/jvi.00344-25. Epub 2025 Jun 2. J Virol. 2025. PMID: 40454785 Free PMC article.
-
Coordination of oxysterol binding protein 1 and VAP-A/B modulates the generation of cholesterol and viral inclusion bodies to promote grass carp reovirus replication.Front Immunol. 2024 Jul 16;15:1419321. doi: 10.3389/fimmu.2024.1419321. eCollection 2024. Front Immunol. 2024. PMID: 39081319 Free PMC article.
-
Grass Carp Reovirus VP56 Allies VP4, Recruits, Blocks, and Degrades RIG-I to More Effectively Attenuate IFN Responses and Facilitate Viral Evasion.Microbiol Spectr. 2021 Oct 31;9(2):e0100021. doi: 10.1128/Spectrum.01000-21. Epub 2021 Sep 15. Microbiol Spectr. 2021. PMID: 34523975 Free PMC article.
Cited by
-
Grass carp Trim47 restricts GCRV infection via SPRY domain-mediated autophagic degradation of nonstructural proteins and disruption of viral inclusion bodies.Front Immunol. 2025 Jul 10;16:1623014. doi: 10.3389/fimmu.2025.1623014. eCollection 2025. Front Immunol. 2025. PMID: 40709172 Free PMC article.
References
-
- Fisheries Bureau of Ministry of Agriculture and Rural Affairs . 2024. China fishery statistical yearbook of 2023. Beijing, China: China Agriculture Press
-
- Wang X, Zhang F, Su R, Li X, Chen W, Chen Q, Yang T, Wang J, Liu H, Fang Q, Cheng L. 2018. Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly. Proc Natl Acad Sci USA 115:7344–7349. doi:10.1073/pnas.1803885115 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

