Autophagy-related protein Atg11 is essential for microtubule-mediated chromosome segregation
- PMID: 40173187
- PMCID: PMC11984983
- DOI: 10.1371/journal.pbio.3003069
Autophagy-related protein Atg11 is essential for microtubule-mediated chromosome segregation
Abstract
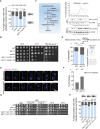
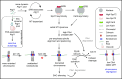
Emerging studies hint at the roles of autophagy-related proteins in various cellular processes. To understand if autophagy-related proteins influence genome stability, we sought to examine a cohort of 35 autophagy mutants in Saccharomyces cerevisiae. We observe cells lacking Atg11 show poor mitotic stability of minichromosomes. Single-molecule tracking assays and live cell microscopy reveal that Atg11 molecules dynamically localize to the spindle pole bodies (SPBs) in a microtubule (MT)-dependent manner. Loss of Atg11 leads to a delayed cell cycle progression. Such cells accumulate at metaphase at an elevated temperature that is relieved when the spindle assembly checkpoint (SAC) is inactivated. Indeed, atg11∆ cells have stabilized securin levels, that prevent anaphase onset. Ipl1-mediated activation of SAC also confirms that atg11∆ mutants are defective in chromosome biorientation. Atg11 functions in the Kar9-dependent spindle positioning pathway. Stabilized Clb4 levels in atg11∆ cells suggest that Atg11 maintains Kar9 asymmetry by facilitating proper dynamic instability of astral microtubules (aMTs). Loss of Spc72 asymmetry contributes to non-random SPB inheritance in atg11∆ cells. Overall, this study uncovers an essential non-canonical role of Atg11 in the MT-mediated process of chromosome segregation.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Byers B. Cytology of the yeast life cycle. Cold Spring Harbor Monogr Arch. 1981;11.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

