Fibroblast atlas: Shared and specific cell types across tissues
- PMID: 40173240
- PMCID: PMC11963979
- DOI: 10.1126/sciadv.ado0173
Fibroblast atlas: Shared and specific cell types across tissues
Abstract
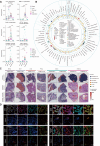
Understanding the heterogeneity of fibroblasts depends on decoding the complexity of cell subtypes, their origin, distribution, and interactions with other cells. Here, we integrated 249,156 fibroblasts from 73 studies across 10 tissues to present a single-cell atlas of fibroblasts. We provided a high-resolution classification of 18 fibroblast subtypes. In particular, we revealed a previously undescribed cell population, TSPAN8+ chromatin remodeling fibroblasts, characterized by high expression of genes with functions related to histone modification and chromatin remodeling. Moreover, TSPAN8+ chromatin remodeling fibroblasts were detectable in spatial transcriptome data and multiplexed immunofluorescence assays. Compared with other fibroblast subtypes, TSPAN8+ chromatin remodeling fibroblasts exhibited higher scores in cell differentiation and resident fibroblast, mainly interacting with endothelial cells and T cells through ligand VEGFA and receptor F2R, and their presence was associated with poor prognosis. Our analyses comprehensively defined the shared and specific characteristics of fibroblast subtypes across tissues and provided a user-friendly data portal, Fibroblast Atlas.
Figures






References
-
- Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., Bassez A., Decaluwe H., Pircher A., Van den Eynde K., Weynand B., Verbeken E., De Leyn P., Liston A., Vansteenkiste J., Carmeliet P., Aerts S., Thienpont B., Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018). - PubMed
-
- Ohlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A. S., Ponz-Sarvise M., Corbo V., Oni T. E., Hearn S. A., Lee E. J., Chio I. I., Hwang C. I., Tiriac H., Baker L. A., Engle D. D., Feig C., Kultti A., Egeblad M., Fearon D. T., Crawford J. M., Clevers H., Park Y., Tuveson D. A., Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017). - PMC - PubMed
-
- Elyada E., Bolisetty M., Laise P., Flynn W. F., Courtois E. T., Burkhart R. A., Teinor J. A., Belleau P., Biffi G., Lucito M. S., Sivajothi S., Armstrong T. D., Engle D. D., Yu K. H., Hao Y., Wolfgang C. L., Park Y., Preall J., Jaffee E. M., Califano A., Robson P., Tuveson D. A., Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 9, 1102–1123 (2019). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

