The Genetic Architecture of Congenital Diarrhea and Enteropathy
- PMID: 40174224
- PMCID: PMC11968080
- DOI: 10.1056/NEJMoa2405333
The Genetic Architecture of Congenital Diarrhea and Enteropathy
Abstract
Background: Next-generation sequencing has enabled precision therapeutic approaches that have improved the lives of children with rare diseases. Congenital diarrhea and enteropathies (CODEs) are associated with high morbidity and mortality. Although treatment of these disorders is largely supportive, emerging targeted therapies based on genetic diagnoses include specific diets, pharmacologic treatments, and surgical interventions.
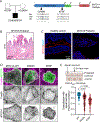
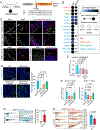
Methods: We analyzed the exomes or genomes of infants with suspected monogenic congenital diarrheal disorders. Using cell and zebrafish models, we tested the effects of variants in newly implicated genes.
Results: In our case series of 129 infant probands with suspected monogenic congenital diarrheal disorders, we identified causal variants, including a new founder NEUROG3 variant, in 62 infants (48%). Using cell and zebrafish models, we also uncovered and functionally characterized three novel genes associated with CODEs: GRWD1, MYO1A, and MON1A.
Conclusions: We have characterized the broad genetic architecture of CODE disorders in a large case series of patients and identified three novel genes associated with CODEs. (Funded by the National Institutes of Health and others.).
Copyright © 2025 Massachusetts Medical Society.
Figures
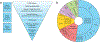

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
