Inhibition of tau neuronal internalization using anti-tau single domain antibodies
- PMID: 40175345
- PMCID: PMC11965310
- DOI: 10.1038/s41467-025-58383-4
Inhibition of tau neuronal internalization using anti-tau single domain antibodies
Abstract
In Alzheimer's disease, tau pathology spreads across brain regions as the disease progresses. Intracellular tau can be released and taken up by nearby neurons. We evaluated single domain anti-tau antibodies, also called VHHs, as inhibitors of tau internalization. We identified three VHH inhibitors of tau uptake: A31, H3-2, and Z70mut1. These VHHs compete with the membrane protein LRP1, a major receptor mediating neuronal uptake of tau. A31 and Z70mut1 bind to microtubule binding domain repeats, which are involved in the interaction with LRP1. VHH H3-2 is the only VHH from our library that reduces the internalization of both monomeric tau and tau fibrils. VHH H3-2 binds a C-terminal tau epitope with high affinity. Its three-dimensional structure in complex with a tau peptide reveals a unique binding mode as a VHH-swapped dimer. These anti-tau VHHs are interesting tools to study tau prion-like propagation in tauopathies and potentially develop novel biotherapies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.-C.R. is the CEO of Hybrigenic Services. C.D., E.D., J.-C. R., L.B., and I.L. are the inventors of a patent (WO2020/120644A1) that covers the use of VHH Z70 and VHHs derived from it. The remaining authors declare no competing interests.
Figures


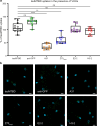

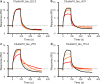


References
-
- Neupane, K. et al. Direct observation of prion-like propagation of protein misfolding templated by pathogenic mutants. Nat. Chem. Biol.20, 1220–1226 (2024). - PubMed
MeSH terms
Substances
Grants and funding
- ANR-11-LABX-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-18-CE44-0016/Agence Nationale de la Recherche (French National Research Agency)
- ANR-22-CE92-0061/Agence Nationale de la Recherche (French National Research Agency)
- GA101119596/European Commission (EC)
- 6433/Association France Alzheimer (French Alzheimer's Association)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

