A unique mechanism of snRNP core assembly
- PMID: 40175367
- PMCID: PMC11965559
- DOI: 10.1038/s41467-025-58461-7
A unique mechanism of snRNP core assembly
Abstract
The assembly of most spliceosomal snRNP cores involves seven Sm proteins (D1/D2/F/E/G/D3/B) forming a ring around snRNA, typically requiring essential assembly chaperones like the SMN complex, associated with spinal muscular atrophy (SMA). Strikingly, in budding yeast, snRNP core assembly only involves Brr1, a nonessential homolog of Gemin2. Here, we reveal two distinct pathways in budding yeast: an inefficient chaperone-mediated pathway involving Brr1 and a novel factor, Lot5, and a direct pathway. Lot5 binds D1/D2/F/E/G to form a heterohexameric ring (6S). Brr1 binds D1/D2/F/E/G and 6S but cannot displace Lot5 to facilitate assembly. Disruption of BRR1 and LOT5 genes caused mild growth retardation, but LOT5 overexpression substantially impeded growth. The direct pathway uniquely involves F/E/G as a trimer and a stable D1/D2/F/E/G intermediate complex, explaining the non-essentiality of chaperones. These findings unveil a unique snRNP core assembly mechanism, illuminate the evolution of assembly chaperones, and suggest avenues for studying SMA pathophysiology.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






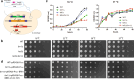

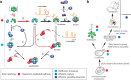
References
-
- Kambach, C. et al. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell96, 375–387 (1999). - PubMed
-
- Chari, A. et al. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell135, 497–509 (2008). - PubMed
MeSH terms
Substances
Grants and funding
- 2017YFA0504300/Ministry of Science and Technology of the People's Republic of China (Chinese Ministry of Science and Technology)
- 2017YFA0505900/Ministry of Science and Technology of the People's Republic of China (Chinese Ministry of Science and Technology)
- 2020YJ0209/Department of Science and Technology of Sichuan Province (Sichuan Provincial Department of Science and Technology)
LinkOut - more resources
Full Text Sources

