Amyloid-reoriented enzyme catalysis
- PMID: 40175427
- PMCID: PMC11965455
- DOI: 10.1038/s41467-025-58536-5
Amyloid-reoriented enzyme catalysis
Abstract
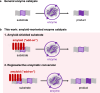
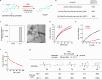
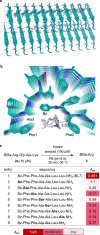
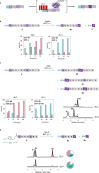
Enzyme catalysis is essential for molecular transformations. Here, we make use of amyloid, a fibrillar aggregate formed by stacking peptides with β-sheet, which offers unique selectivity in enzymatic reactions. Azo-stilbene derivative (ASB), the amyloid-recognition motif, is incorporated into the substrate, which allows the amyloid consisting of Bz-Phe-Phe-Ala-Ala-Leu-Leu-NH2 (BL7) to shield the substrates from the approaching enzyme. X-ray crystallographic analysis and structure-shielding effect relationship studies of BL7 reveal that the benzene rings present in the N-terminal benzoyl group and Phe1 side chain are particularly important for the shielding effect on the substrate. The finding results in a selective transformation system in which the reactive site close to ASB is protected by amyloid, while a site far from ASB is converted by the enzymes (trypsin, protein arginine deiminase [PAD], and Staphylococcus aureus V-8 Protease [Glu-C]). Further, the amyloid-shielded enzyme catalysis is compatible with an intact peptide, as the side chain of Tyr can be converted to the amyloid-recognizing motif. The enzymatic reactions combining amyloid provide unique selectivity for molecular transformation which may be used in diverse fields, including in synthetic chemistry.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Chen, K. & Arnold, F. H. Engineering new catalytic activities in enzymes. Nat. Catal.3, 203–213 (2020).
-
- Wang, Y. et al. Directed evolution: methodologies and applications. Chem. Rev.121, 12384–12444 (2021). - PubMed
-
- Zhang, X. et al. Enzymatic synthesis of organoselenium compounds via C‒Se bond formation mediated by sulfur carrier proteins. Nat. Synth.3, 477–487 (2024).
-
- Huffman, M. A. et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science366, 1255–1259 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- JP23K14325/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP24K02153/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP24H01787/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP24K09344/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP23ama121001/Japan Agency for Medical Research and Development (AMED)
LinkOut - more resources
Full Text Sources

