Ancient DNA from the Green Sahara reveals ancestral North African lineage
- PMID: 40175549
- PMCID: PMC12043513
- DOI: 10.1038/s41586-025-08793-7
Ancient DNA from the Green Sahara reveals ancestral North African lineage
Abstract
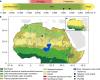
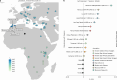
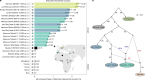
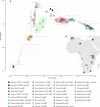
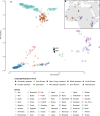
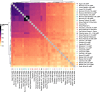
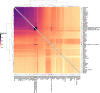
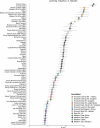
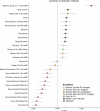
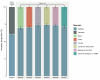
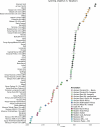
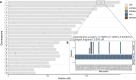
Although it is one of the most arid regions today, the Sahara Desert was a green savannah during the African Humid Period (AHP) between 14,500 and 5,000 years before present, with water bodies promoting human occupation and the spread of pastoralism in the middle Holocene epoch1. DNA rarely preserves well in this region, limiting knowledge of the Sahara's genetic history and demographic past. Here we report ancient genomic data from the Central Sahara, obtained from two approximately 7,000-year-old Pastoral Neolithic female individuals buried in the Takarkori rock shelter in southwestern Libya. The majority of Takarkori individuals' ancestry stems from a previously unknown North African genetic lineage that diverged from sub-Saharan African lineages around the same time as present-day humans outside Africa and remained isolated throughout most of its existence. Both Takarkori individuals are closely related to ancestry first documented in 15,000-year-old foragers from Taforalt Cave, Morocco2, associated with the Iberomaurusian lithic industry and predating the AHP. Takarkori and Iberomaurusian-associated individuals are equally distantly related to sub-Saharan lineages, suggesting limited gene flow from sub-Saharan to Northern Africa during the AHP. In contrast to Taforalt individuals, who have half the Neanderthal admixture of non-Africans, Takarkori shows ten times less Neanderthal ancestry than Levantine farmers, yet significantly more than contemporary sub-Saharan genomes. Our findings suggest that pastoralism spread through cultural diffusion into a deeply divergent, isolated North African lineage that had probably been widespread in Northern Africa during the late Pleistocene epoch.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations.Science. 2018 May 4;360(6388):548-552. doi: 10.1126/science.aar8380. Epub 2018 Mar 15. Science. 2018. PMID: 29545507
-
Ancient DNA and deep population structure in sub-Saharan African foragers.Nature. 2022 Mar;603(7900):290-296. doi: 10.1038/s41586-022-04430-9. Epub 2022 Feb 23. Nature. 2022. PMID: 35197631 Free PMC article.
-
Genomic ancestry of North Africans supports back-to-Africa migrations.PLoS Genet. 2012 Jan;8(1):e1002397. doi: 10.1371/journal.pgen.1002397. Epub 2012 Jan 12. PLoS Genet. 2012. PMID: 22253600 Free PMC article.
-
African human mtDNA phylogeography at-a-glance.J Anthropol Sci. 2011;89:25-58. doi: 10.4436/jass.89006. Epub 2011 Mar 15. J Anthropol Sci. 2011. PMID: 21368343 Review.
-
Population history of North Africa based on modern and ancient genomes.Hum Mol Genet. 2021 Apr 26;30(R1):R17-R23. doi: 10.1093/hmg/ddaa261. Hum Mol Genet. 2021. PMID: 33284971 Review.
Cited by
-
The origin of modern North Africans as depicted by a massive survey of mitogenomes.Sci Rep. 2025 Jul 25;15(1):27025. doi: 10.1038/s41598-025-12209-x. Sci Rep. 2025. PMID: 40707575 Free PMC article.
References
-
- Kuper, R. & Kröpelin, S. Climate-controlled Holocene occupation in the Sahara: motor of Africa’s evolution. Science313, 803–807 (2006). - PubMed
-
- van de Loosdrecht, M. et al. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science360, 548–552 (2018). - PubMed
-
- COHMAP Members. Climatic changes of the last 18,000 years: observations and model simulations. Science241, 1043–1052 (1988). - PubMed
-
- Ritchie, J. C., Eyles, C. H. & Haynes, C. V. Sediment and pollen evidence for an early to mid-Holocene humid period in the eastern Sahara. Nature314, 352–355 (1985).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

