Molecular characterization of virulent genes in Pseudomonas aeruginosa based on componential usage divergence
- PMID: 40175567
- PMCID: PMC11965391
- DOI: 10.1038/s41598-025-95579-6
Molecular characterization of virulent genes in Pseudomonas aeruginosa based on componential usage divergence
Abstract
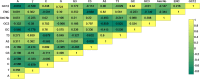
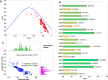
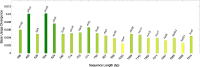
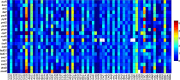
Genetic characteristics of virulent genes in Pseudomonas aeruginosa attracted significant attention for they could govern their drug-resistances. Studies on the componential usage divergences in the virulent genes are beneficial for further explicating their molecular characteristics. In present study, one thousand complete genomes of Pseudomonas aeruginosa were considered to study the molecular characteristics of 21 typical virulent genes. The important componential usage patterns (i.e., the base usage pattern, the codon usage pattern and their divergences) of 21 specific virulent genes were counted and calculated. The results show that (1) most virulent genes concerned in the present study are high GC sequences (overall GC ratio > 50%), especially from the codon usage perspective, the virulent genes are obviously GC3-abundant sequences (GC3 ratio > 70%); (2) the relative synonymous codon usage of all concerned virulent genes are uneven, especially in the anvM and the lptA, there is no codon for some certain amino acids, which could reveal their obvious codon usage bias; (3) some genes (i.e., the oprF and the fadD1) with lower divergence have steady effective number of codons. The findings of the present work would improve novel insights on the genetic characteristics of virulent genes in Pseudomonas aeruginosa.
Keywords: Pseudomonas aeruginosa; Biodiversity; Codon usage bias; Evolutionary pressure; Virulent gene.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Codon usage divergence of important functional genes in Mycobacterium tuberculosis.Int J Biol Macromol. 2022 Jun 1;209(Pt A):1197-1204. doi: 10.1016/j.ijbiomac.2022.04.112. Epub 2022 Apr 21. Int J Biol Macromol. 2022. PMID: 35460756
-
Synonymous codon usage in Pseudomonas aeruginosa PA01.Gene. 2002 May 1;289(1-2):131-9. doi: 10.1016/s0378-1119(02)00503-6. Gene. 2002. PMID: 12036591
-
Codon usage divergence in Delta variants (B.1.617.2) of SARS-CoV-2.Infect Genet Evol. 2022 Jan;97:105175. doi: 10.1016/j.meegid.2021.105175. Epub 2021 Dec 3. Infect Genet Evol. 2022. PMID: 34871776 Free PMC article.
-
Codon usage and codon pair patterns in non-grass monocot genomes.Ann Bot. 2017 Nov 28;120(6):893-909. doi: 10.1093/aob/mcx112. Ann Bot. 2017. PMID: 29155926 Free PMC article. Review.
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
References
-
- Gellatly, S. L. & Hancock, R. E. W. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis.67, 159–173. 10.1111/2049-632X.12033 (2013). - PubMed
-
- Botelho, J., Grosso, F. & Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa - Mechanisms, epidemiology and evolution. Drug Resist. Updates44, 26–47. 10.1016/j.drup.2019.07.002 (2019). - PubMed
-
- Jault, P. et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis.19, 35–45. 10.1016/S1473-3099(18)30482-1 (2019). - PubMed
-
- Pang, Z., Raudonis, R., Glick, B. R., Lin, T. & Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv.37, 177–192. 10.1016/j.biotechadv.2018.11.013 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

