Requirement for Cyclin D1 Underlies Cell-Autonomous HIF2 Dependence in Kidney Cancer
- PMID: 40178040
- PMCID: PMC12223508
- DOI: 10.1158/2159-8290.CD-24-1378
Requirement for Cyclin D1 Underlies Cell-Autonomous HIF2 Dependence in Kidney Cancer
Abstract
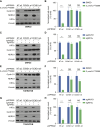
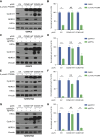
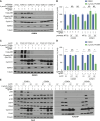
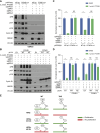
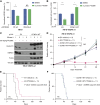
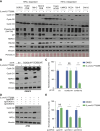
Inactivation of the VHL gene stabilizes HIF2α, which drives clear-cell renal cell carcinoma (ccRCC). The HIF2α inhibitor belzutifan is approved for ccRCC treatment, but de novo and acquired resistance are common. HIF2α, bound to ARNT, transcriptionally activates many genes. We performed CRISPR-mediated gene activation screens in HIF2α-dependent ccRCC lines treated with a belzutifan analog to identify HIF2α-responsive genes that confer cell-autonomous belzutifan resistance when not downregulated. Sustaining the expression of the HIF2α target gene CCND1, encoding cyclin D1, promoted HIF2α independence/belzutifan resistance. This activity requires CDK4/6 activation by cyclin D1 but is not solely due to phosphorylation of the canonical cyclin D1 target, pRB. Indeed, ccRCC lines lacking all three pRB family members remained at least partially HIF2α-dependent. In this context, however, a kinase-defective cyclin D1 variant partially overrode belzutifan's antiproliferative effects, suggesting that ccRCC promotion by cyclin D1 requires the phosphorylation of pRB paralogs and one or more kinase-independent cyclin D1 activities.
Significance: We discovered that cyclin D1 is the key target of HIF2 driving the cell-autonomous proliferation of VHL-mutant kidney cancers and that cyclin D1 has targets beyond pRB in this setting. These findings have implications for treating kidney cancer with HIF2 inhibitors, alone or in combination with CDK4/6 inhibitors.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
J.G. Doench reports personal fees from Pfizer, Microsoft Research, BioNTech, and from Tango Therapeutics outside the submitted work. W.R. Sellers reports personal fees from Peleton Therapeutics and grants from Merck Pharmaceuticals during the conduct of the study, as well as grants from Pfizer Pharmaceuticals, Bristol Myers Squibb, Takeda Pharmaceuticals, Boehringer-Ingelheim, Calico Biosciences, and Ridgeline Discovery and personal fees from Redridge Bio and Delphia Therapeutics outside the submitted work. W.G. Kaelin reports grants from the NIH and grants and other support from Howard Hughes Medical Institute during the conduct of the study, as well as nonfinancial support and other support from Merck (including Belzutifan milestone payments), personal fees and other support from Nextech Invest, Lilly Pharmaceuticals, LifeMine Therapeutics, Circle Pharmaceuticals, and Tango Therapeutics, and personal fees from Casdin Capital, Arcus Biosciences, Red Tree Ventures, Fibrogen, and Regeneron Ventures outside the submitted work. No disclosures were reported by the other authors.
Figures

References
-
- Maher ER, Kaelin WG Jr. von Hippel-Lindau disease. Medicine (Baltimore) 1997;76:381–91. - PubMed
-
- Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol 2004;22:4991–5004. - PubMed
-
- Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013;45:860–7. - PubMed
-
- Choueiri TK, Kaelin WG Jr. Targeting the HIF2-VEGF axis in renal cell carcinoma. Nat Med 2020;26:1519–30. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

