Polyyne production is regulated by the transcriptional regulators PgnC and GacA in Pseudomonas protegens Pf-5
- PMID: 40178257
- PMCID: PMC12016544
- DOI: 10.1128/aem.02388-24
Polyyne production is regulated by the transcriptional regulators PgnC and GacA in Pseudomonas protegens Pf-5
Abstract
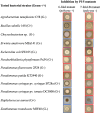
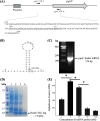
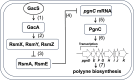
Polyynes produced by bacteria have promising applications in agriculture and medicine due to their potent antimicrobial activities. Polyyne biosynthetic genes have been identified in Pseudomonas and Burkholderia. However, the molecular mechanisms underlying the regulation of polyyne biosynthesis remain largely unknown. In this study, we used a soil bacterium Pseudomonas protegens Pf-5, which was recently reported to produce polyyne called protegenin, as a model to investigate the regulation of bacterial polyyne production. Our results show that Pf-5 controls polyyne production at both the pathway-specific level and a higher global level. Mutation of pgnC, a transcriptional regulatory gene located in the polyyne biosynthetic gene cluster, abolished polyyne production. Gene expression analysis revealed that PgnC directly activates the promoter of polyyne biosynthetic genes. The production of polyyne also requires a global regulator GacA. Mutation of gacA decreased the translation of PgnC, which is consistent with the result that pgnC leader mRNA bound directly to RsmE, an RNA-binding protein negatively regulated by GacA. These results suggest that GacA induces the expression of the PgnC regulator, which in turn activates polyyne biosynthesis. Additionally, the polyyne-producing strain of Pf-5, but not the polyyne-nonproducing strain, could inhibit a broad spectrum of bacteria including both Gram-negative and Gram-positive bacteria.IMPORTANCEAntimicrobial metabolites produced by bacteria are widely used in agriculture and medicine to control plant, animal, and human pathogens. Although bacteria-derived polyynes have been identified as potent antimicrobials for decades, the molecular mechanisms by which bacteria regulate polyyne biosynthesis remain understudied. In this study, we found that polyyne biosynthesis is directly activated by a pathway-specific regulator PgnC, which is induced by a global regulator GacA through the RNA-binding protein RsmE in Pseudomonas protegens. To our knowledge, this work is the first comprehensive study of the regulatory mechanisms of bacterial polyyne biosynthesis at both pathway-specific level and global level. The discovered molecular mechanisms can help us optimize polyyne production for agricultural or medical applications.
Keywords: Pseudomonas protegens; antimicrobial activity; gene regulation; polyyne biosynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Keswani C, Singh HB, García-Estrada C, Caradus J, He Y-W, Mezaache-Aichour S, Glare TR, Borriss R, Sansinenea E. 2020. Antimicrobial secondary metabolites from agriculturally important bacteria as next-generation pesticides. Appl Microbiol Biotechnol 104:1013–1034. doi:10.1007/s00253-019-10300-8 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

