Comparative Analysis of Drosophila Bam and Bgcn Sequences and Predicted Protein Structural Evolution
- PMID: 40178596
- PMCID: PMC12006264
- DOI: 10.1007/s00239-025-10245-9
Comparative Analysis of Drosophila Bam and Bgcn Sequences and Predicted Protein Structural Evolution
Abstract
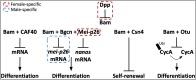
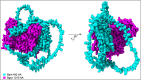
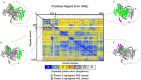
The protein encoded by the Drosophila melanogaster gene bag of marbles (bam) plays an essential role in early gametogenesis by complexing with the gene product of benign gonial cell neoplasm (bgcn) to promote germline stem cell daughter differentiation in males and females. Here, we compared the AlphaFold2 and AlphaFold Multimer predicted structures of Bam protein and the Bam:Bgcn protein complex between D. melanogaster, D. simulans, and D. yakuba, where bam is necessary in gametogenesis to that in D. teissieri, where it is not. Despite significant sequence divergence, we find very little evidence of significant structural differences in high confidence regions of the structures across the four species. This suggests that Bam structure is unlikely to be a direct cause of its functional differences between species and that Bam may simply not be integrated in an essential manner for GSC differentiation in D. teissieri. Patterns of positive selection and significant amino acid diversification across species is consistent with the Selection, Pleiotropy, and Compensation (SPC) model, where detected selection at bam is consistent with adaptive change in one major trait followed by positively selected compensatory changes for pleiotropic effects (in this case perhaps preserving structure). In the case of bam, we suggest that the major trait could be genetic interaction with the endosymbiotic bacteria Wolbachia pipientis. Following up on detected signals of positive selection and comparative structural analysis could provide insight into the distribution of a primary adaptive change versus compensatory changes following a primary change.
Keywords: Bam; Bgcn; AlphaFold; Orthologs; Protein structure; Reproduction.
© 2025. The Author(s).
Figures





Update of
-
Comparative Analysis of Drosophila Bam and Bgcn Sequences and Predicted Protein Structural Evolution.bioRxiv [Preprint]. 2024 Dec 18:2024.12.17.628990. doi: 10.1101/2024.12.17.628990. bioRxiv. 2024. Update in: J Mol Evol. 2025 Apr;93(2):278-291. doi: 10.1007/s00239-025-10245-9. PMID: 39763920 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

