Dissection of single-cell landscapes for the development of chimeric antigen receptor T cells in Hodgkin lymphoma
- PMID: 40178843
- PMCID: PMC12002222
- DOI: 10.1182/blood.2023022197
Dissection of single-cell landscapes for the development of chimeric antigen receptor T cells in Hodgkin lymphoma
Abstract
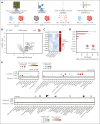
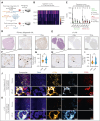
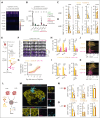
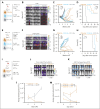
The success of targeted therapies for hematological malignancies has heralded their potential as both salvage treatment and early treatment lines, reducing the need for high-dose, intensive, and often toxic chemotherapeutic regimens. For young patients with classic Hodgkin lymphoma (cHL), immunotherapies provide the possibility to lessen long-term, treatment-related toxicities. However, suitable therapeutic targets are lacking. By integrating single-cell dissection of the tumor landscape and an in-depth, single-cell-based off-tumor antigen prediction, we identify CD86 as a promising therapeutic target in cHL. CD86 is highly expressed on Hodgkin and Reed-Sternberg cancer cells and cHL-specific tumor-associated macrophages. We reveal CD86-CTLA-4 as a key suppressive pathway in cHL, driving T-cell exhaustion. Cellular therapies targeting CD86 had extraordinary efficacy in vitro and in vivo and were safe in immunocompetent mouse models without compromising bacterial host defense in sepsis models. Our results prove the potential value of anti-CD86 immunotherapies for treating cHL.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: A.G., M. Thomas, R.G., S.E., C.M., and S.K. submitted patent applications related to this work filed by Ludwig Maximilians Universität München, the University Hospital of LMU Munich, or the Helmholtz Centre Munich. S.K. has received honoraria from Cymab, Plectonic, TCR2 Inc, Novartis, Bristol Myers Squibb, Miltenyi, and GlaxoSmithKline. S.K. and S.E. are inventors of several patents in the field of immunooncology. S.K. and S.E. received license fees from TCR2 Inc and Carina Biotech. A.G. received research support from Tabby Therapeutics and Plectonic for work unrelated to the manuscript. P.J.B. reports advisory roles for Hexal, Merck Sharp & Dohme, Need Inc, Stemline, and Takeda; holds stock options in Need Inc; has received honoraria from AstraZeneca, BeiGene, Bristol Myers Squibb/Celgene, Lilly, Merck Sharp & Dohme, Need Inc, Stemline, and Takeda; and has received research funding from BeiGene (Institution), Bristol Myers Squibb (Institution), Merck Sharp & Dohme (Institution), and Takeda (Institution); all unrelated to the manuscript. S.K. and S.E. received research support from TCR2 Inc, Plectonic GmBH, CatalYm GmBH, and Arcus Bioscience for work unrelated to the manuscript. S.F.-W. and C.K. are inventors on a patent application covering bone marrow organoids (WO2023/194370A1). The remaining authors declare no competing financial interests.
Figures




References
-
- Majzner RG, Mackall CL. Clinical lessons learned from the first leg of the CAR T cell journey. Nat Med. 2019;25(9):1341–1355. - PubMed
-
- Labrijn AF, Janmaat ML, Reichert JM, Parren P. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. 2019;18(8):585–608. - PubMed
-
- Bishop MR, Dickinson M, Purtill D, et al. Tisagenlecleucel vs standard of care as second-line therapy of primary refractory or relapsed aggressive B-cell non-Hodgkin lymphoma: analysis of the phase III Belinda study [abstract] Blood. 2021;138(suppl 2):LBA-6.
-
- Locke FL, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel as second-line therapy for large B-cell lymphoma. N Engl J Med. 2022;386(7):640–654. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

