Hyaluronan network remodeling by ZEB1 and ITIH2 enhances the motility and invasiveness of cancer cells
- PMID: 40178908
- PMCID: PMC12126249
- DOI: 10.1172/JCI180570
Hyaluronan network remodeling by ZEB1 and ITIH2 enhances the motility and invasiveness of cancer cells
Abstract
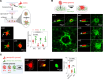
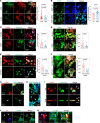
Hyaluronan (HA) in the extracellular matrix promotes epithelial-mesenchymal transition (EMT) and metastasis; however, the mechanism by which the HA network constructed by cancer cells regulates cancer progression and metastasis in the tumor microenvironment (TME) remains largely unknown. In this study, inter-α-trypsin inhibitor heavy chain 2 (ITIH2), an HA-binding protein, was confirmed to be secreted from mesenchymal-like lung cancer cells when cocultured with cancer-associated fibroblasts. ITIH2 expression is transcriptionally upregulated by the EMT-inducing transcription factor ZEB1, along with HA synthase 2 (HAS2), which positively correlates with ZEB1 expression. Depletion of ITIH2 and HAS2 reduced HA matrix formation and the migration and invasion of lung cancer cells. Furthermore, ZEB1 facilitates alternative splicing and isoform expression of CD44, an HA receptor, and CD44 knockdown suppresses the motility and invasiveness of lung cancer cells. Using a deep learning-based drug-target interaction algorithm, we identified an ITIH2 inhibitor (sincalide) that inhibited HA matrix formation and migration of lung cancer cells, preventing metastatic colonization of lung cancer cells in mouse models. These findings suggest that ZEB1 remodels the HA network in the TME through the regulation of ITIH2, HAS2, and CD44, presenting a strategy for targeting this network to suppress lung cancer progression.
Keywords: Cell biology; Extracellular matrix; Lung cancer; Oncology.
Conflict of interest statement
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

