Dipeptidase-1-knockout mice develop invasive tumors with features of microsatellite-unstable colorectal cancer
- PMID: 40178918
- PMCID: PMC12128987
- DOI: 10.1172/jci.insight.186938
Dipeptidase-1-knockout mice develop invasive tumors with features of microsatellite-unstable colorectal cancer
Abstract
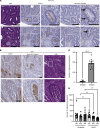
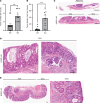
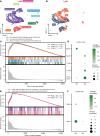
Dipeptidase-1 (DPEP1) is highly upregulated in colorectal cancer (CRC), with its enzymatic function linked to invasion and metastasis. More recently, DPEP1 was found to serve as a receptor for neutrophils when expressed by activated endothelial cells. It is unknown whether neutrophils bind to DPEP1-expressing CRC cells and whether this impacts features of CRC. Neutrophils have been shown to be tumor promoting in cancers including CRC, where they act to exclude CD8+ T cells. Herein, we show that neutrophils bind DPEP1-expressing CRC cells. In addition, DPEP1 is preferentially expressed in microsatellite-stable (MSS) CRCs, in which there are a paucity of CD8+ T cells, whereas DPEP1 is negatively correlated with microsatellite-unstable (MSI-H) CRCs, which are T cell rich and are more responsive to immunotherapy. Remarkably, carcinogen-treated Dpep1-null mice develop multiple, large, plaque-like, locally invasive adenocarcinomas and squamous cell cancers in the distal colon. These adenocarcinomas exhibit a marked reduction in neutrophils and an influx CD8+ T cells, along with reduced expression of mismatch repair proteins, consistent with features of MSI-H CRC. These results establish DPEP1's importance in maintaining MSS CRC and its ability to shape the tumor microenvironment.
Keywords: Cancer; Cell biology; Oncology.
Conflict of interest statement
Figures



References
-
- Buckhaults P, et al. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61(19):6996–7001. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

