Playbook workflow builder: Interactive construction of bioinformatics workflows
- PMID: 40179105
- PMCID: PMC11967941
- DOI: 10.1371/journal.pcbi.1012901
Playbook workflow builder: Interactive construction of bioinformatics workflows
Abstract
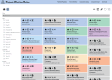
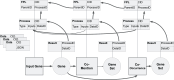
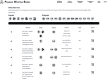
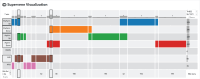
The Playbook Workflow Builder (PWB) is a web-based platform to dynamically construct and execute bioinformatics workflows by utilizing a growing network of input datasets, semantically annotated API endpoints, and data visualization tools contributed by an ecosystem of collaborators. Via a user-friendly user interface, workflows can be constructed from contributed building-blocks without technical expertise. The output of each step of the workflow is added into reports containing textual descriptions, figures, tables, and references. To construct workflows, users can click on cards that represent each step in a workflow, or construct workflows via a chat interface that is assisted by a large language model (LLM). Completed workflows are compatible with Common Workflow Language (CWL) and can be published as research publications, slideshows, and posters. To demonstrate how the PWB generates meaningful hypotheses that draw knowledge from across multiple resources, we present several use cases. For example, one of these use cases prioritizes drug targets for individual cancer patients using data from the NIH Common Fund programs GTEx, LINCS, Metabolomics, GlyGen, and ExRNA. The workflows created with PWB can be repurposed to tackle similar use cases using different inputs. The PWB platform is available from: https://playbook-workflow-builder.cloud/.
Copyright: © 2025 Clarke et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Cohen-Boulakia S, Belhajjame K, Collin O, Chopard J, Froidevaux C, Gaignard A, et al.. Scientific workflows for computational reproducibility in the life sciences: Status, challenges and opportunities. Future Gener Comput Syst. 2017;75:284–98. doi: 10.1016/j.future.2017.01.012 - DOI

