Metagenomic Identification of Fusarium solani Strain as Cause of US Fungal Meningitis Outbreak Associated with Surgical Procedures in Mexico, 2023
- PMID: 40180580
- PMCID: PMC12044249
- DOI: 10.3201/eid3105.241657
Metagenomic Identification of Fusarium solani Strain as Cause of US Fungal Meningitis Outbreak Associated with Surgical Procedures in Mexico, 2023
Abstract
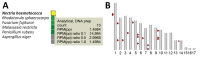
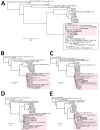
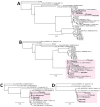
We used metagenomic next-generation sequencing (mNGS) to investigate an outbreak of Fusarium solani meningitis in US patients who had surgical procedures under spinal anesthesia in Matamoros, Mexico, during 2023. Using a novel method called metaMELT (metagenomic multiple extended locus typing), we performed phylogenetic analysis of concatenated mNGS reads from 4 patients (P1-P4) in parallel with reads from 28 fungal reference genomes. Fungal strains from the 4 patients were most closely related to each other and to 2 cultured isolates from P1 and an additional case (P5), suggesting that all cases arose from a point source exposure. Our findings support epidemiologic data implicating a contaminated drug or device used for epidural anesthesia as the likely cause of the outbreak. In addition, our findings show that the benefits of mNGS extend beyond diagnosis of infections to public health outbreak investigation.
Keywords: Fusarium solani; Mexico; Nectria haematococca; SURPI+ computational pipeline for pathogen detection; United States; agnostic pathogen detection; fungi; mNGS; meningitis/encephalitis; metaMELT; metagenomic next-generation sequencing; multiple extended locus typing; outbreak investigation.
Figures

References
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

