Genome-wide CRISPR screen identifies BUB1 kinase as a druggable vulnerability in malignant pleural mesothelioma
- PMID: 40180891
- PMCID: PMC11968822
- DOI: 10.1038/s41419-025-07587-z
Genome-wide CRISPR screen identifies BUB1 kinase as a druggable vulnerability in malignant pleural mesothelioma
Abstract
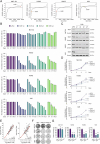
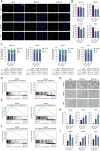
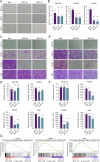
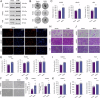
Malignant pleural mesothelioma (MPM) is a rare yet highly aggressive malignancy with a severe prognosis. Compounded by the lack of effective treatment modalities, MPM remains a formidable health challenge. Therefore, the identification of actionable liabilities is critical for advancing precision medicine to combat this lethal disease. Here, we exploit an unbiased genome-wide CRISPR screen, integrating and cross-comparing three MPM cell lines with nonmalignant mesothelial cells, to selectively map the gene targets whose depletion indicates a common dependency in MPM cells. This systematic approach unveils a cohort of verifiable genes, among which BUB1, a mitotic checkpoint serine/threonine kinase, emerges as a high-confidence hit in cancer cells. Cellular and molecular studies demonstrate that genetic depletion or pharmacological inhibition of BUB1 profoundly impairs MPM cell survival and growth while inducing G2/M cell cycle arrest, cellular senescence, and apoptosis, and attenuating functional hallmarks of aggressive cancer cells. Transcriptomic profiling of BUB1-depleted cells discloses differential gene expression signatures congruent with cell fate phenotypes, including the reprogramming of mitotic network genes. Mechanistically, BUB1 is indispensable for the proper localization of essential mitotic regulators MAD1, MAD2, and Shugoshin (SGO1), thereby ensuring the functionality of the spindle assembly checkpoint (SAC). Furthermore, BUB1 ablation leads to cytokinesis failure and multinucleation, a phenotype characterized by the downregulation of CDC20, Cyclin A, and Cyclin B, and a reciprocal upregulation of the cyclin-dependent kinase inhibitor p21. Clinically, MPM tumors exhibit elevated levels of BUB1, and high BUB1 expression is associated with shorter patient survival. Our novel findings accentuate comparative CRISPR screens as a powerful platform to explore tumor cell-selective gene essentiality and propose BUB1 kinase as a potential marker and druggable vulnerability with therapeutic implications for MPM.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: All methods were performed in accordance with the relevant guidelines and regulations.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Scherpereel A, Wallyn F, Albelda SM, Munck C. Novel therapies for malignant pleural mesothelioma. Lancet Oncol. 2018;19:e161–72. - PubMed
-
- Bronte G, Incorvaia L, Rizzo S, Passiglia F, Galvano A, Rizzo F, et al. The resistance related to targeted therapy in malignant pleural mesothelioma: Why has not the target been hit yet? Crit Rev Oncol Hematol. 2016;107:20–32. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

