Integrated proteogenomic characterization of localized prostate cancer identifies biological insights and subtype-specific therapeutic strategies
- PMID: 40180929
- PMCID: PMC11968977
- DOI: 10.1038/s41467-025-58569-w
Integrated proteogenomic characterization of localized prostate cancer identifies biological insights and subtype-specific therapeutic strategies
Abstract
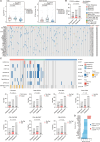
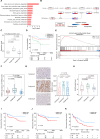
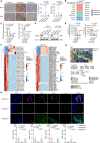
Localized prostate cancer (PCa) is highly variable in their response to therapies. Although a fraction of this heterogeneity can be explained by clinical factors or genomic and transcriptomic profiling, the proteomic-based profiling of aggressive PCa remains poorly understood. Here, we profiled the genome, transcriptome, proteome and phosphoproteome of 145 cases of localized PCa in Chinese patients. Proteome-based stratification of localized PCa revealed three subtypes with distinct molecular features: immune subgroup, arachidonic acid metabolic subgroup and sialic acid metabolic subgroup with highest biochemical recurrence (BCR) rates. Further, we nominated NANS protein, a key enzyme in sialic acid synthesis as a potential prognostic biomarker for aggressive PCa and validated in two independent cohorts. Finally, taking advantage of cell-derived orthotopic transplanted mouse models, single-cell RNA sequencing (scRNA-seq) and immunofluorescence analysis, we revealed that targeting NANS can reverse the immunosuppressive microenvironment through restricting the sialoglycan-sialic acid-recognizing immunoglobulin superfamily lectin (Siglec) axis, thereby inhibiting tumor growth of PCa. In sum, we integrate multi-omic data to refine molecular subtyping of localized PCa, and identify NANS as a potential prognostic biomarker and therapeutic option for aggressive PCa.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer J. Clin.71, 209–249 (2021). - PubMed
-
- Mottet, N. et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol.79, 243–262 (2021). - PubMed
-
- Buyyounouski, M. K., Pickles, T., Kestin, L. L., Allison, R. & Williams, S. G. Validating the interval to biochemical failure for the identification of potentially lethal prostate cancer. J. Clin. Oncol.30, 1857–1863 (2012). - PubMed
-
- Bastian, P. J. et al. High-risk prostate cancer: from definition to contemporary management. Eur. Urol.61, 1096–1106 (2012). - PubMed
-
- Klein, E. A. et al. A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy. Eur. Urol.67, 778–786 (2015). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

