Epigenetic mechanisms controlling human leukemia stem cells and therapy resistance
- PMID: 40180954
- PMCID: PMC11968996
- DOI: 10.1038/s41467-025-58370-9
Epigenetic mechanisms controlling human leukemia stem cells and therapy resistance
Abstract
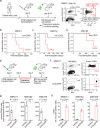
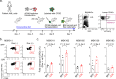
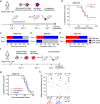
Cancer stem cells are essential for initiation and therapy resistance of many cancers, including acute myeloid leukemias (AML). Here, we apply functional genomic profiling to diverse human leukemias, including high-risk MLL- and NUP98-rearranged specimens, using label tracing in vivo. Human leukemia propagation is mediated by a rare quiescent label-retaining cell (LRC) population undetectable by current immunophenotypic markers. AML quiescence is reversible, preserving genetic clonal competition and epigenetic inheritance. LRC quiescence is defined by distinct promoter-centered chromatin and gene expression dynamics controlled by an AP-1/ETS transcription factor network, where JUN is necessary and sufficient for LRC quiescence and associated with persistence and chemotherapy resistance in diverse patients. This enables prospective isolation and manipulation of immunophenotypically-varied leukemia stem cells, establishing the functions of epigenetic plasticity in leukemia development and therapy resistance. These findings offer insights into leukemia stem cell quiescence and the design of therapeutic strategies for their clinical identification and control.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. A.K. is a consultant to Rgenta, Novartis, Blueprint Medicines, and Syndax.
Figures







References
-
- Lapidot, T. et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature367, 645–648 (1994). - PubMed
-
- Bonnet, D. & Dick, J. E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med.3, 730–737 (1997). - PubMed
-
- Goardon, N. et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell19, 138–152 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

