Peptides developed against receptor binding sites of the E glycoprotein neutralize tick-borne encephalitis virus
- PMID: 40181101
- PMCID: PMC11968884
- DOI: 10.1038/s41598-025-95449-1
Peptides developed against receptor binding sites of the E glycoprotein neutralize tick-borne encephalitis virus
Abstract
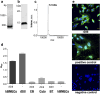
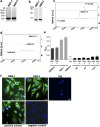
Infection caused by tick-borne encephalitis virus (TBEV) often manifests with meningitis or encephalitis. Domain III (DIII) of the envelope (E) glycoprotein on the TBEV surface facilitates virion attachment to the cell receptor and initiates cell entry. As a result, this research focused on the DIII to develop antiviral molecules that could prevent cell entry. We first identified two receptor-binding sites (RBS, 300SGLTYTMCDK309 and 317APTDSGHDTVVMEVTFSGTKPCR339) on DIII, that are involved in binding to human brain microvascular endothelial cells (hBMECs). Then we sought to isolate peptides from two combinatorial peptide-phage libraries that bind to the RBS and prevent protein E from attaching to hBMECs. Three cyclic peptides (CP2-CNSSKLHMC, CP20-CDGRPDRAC, CP23-CMKESIRGC) and a linear peptide (LP16-AFHPRQMETQMY) inhibited DIII binding to hBMECs. CP2 and CP20, both non-cytotoxic and hemocompatible, effectively neutralized a live virus. CP2 and CP20 deserve further attention for their potential application as anti-TBEV therapeutics that prevent viral cell entry.
Keywords: Blocking peptides; Combinatorial phage library; Domain III; PRNT; Phage display; Tick-borne encephalitis virus; Virus neutralization; hBMECs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

