Hybridization and introgression of the mitochondrial genome between the two species Anisakis pegreffii and A. simplex (s.s.) using a wide genotyping approach: evolutionary and ecological implications
- PMID: 40181623
- PMCID: PMC12186100
- DOI: 10.1017/S0031182025000228
Hybridization and introgression of the mitochondrial genome between the two species Anisakis pegreffii and A. simplex (s.s.) using a wide genotyping approach: evolutionary and ecological implications
Abstract
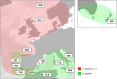
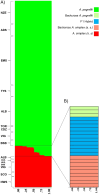
Anisakis pegreffii and A. simplex (s.s.) are the two zoonotic anisakids infecting cetaceans as well as pelagic/demersal fish and squids. In European waters, A. pegreffii prevails in the Mediterranean Sea, while A. simplex (s.s.) in the NE Atlantic Ocean. Abiotic conditions likely play a significant role in shaping their geographical distribution. The Iberian Atlantic and Alboran Sea waters are sympatric areas of the two species. A total of 429 adults and L3 stage from both sympatric and allopatric areas were studied by a wide nuclear genotyping approach (including newly and previously found diagnostic single nucleotide polymorphisms (SNPs) at nuclear DNA (nDNA) and microsatellite DNA loci) and sequenced at mitochondrial DNA (mtDNA) cox2. Admixture between the two species was detected in the sympatric areas studied by STRUCTURE Bayesian analysis; NEWHYBRIDS revealed different categories of hybridization between the two species, representing approximately 5%. A tendency for F1 female hybrids to interbreed with the parental species at the geographical distribution limits of both species was observed. This finding suggests that hybridization occurs when the two parental species significantly differ in abundance. Mitochondrial introgression of A. simplex (s.s.) in A. pegreffii from Mediterranean waters was also detected, likely as a result of past and/or paleo-introgression events. The high level of genetic differentiation between the two species and their backcrosses indicates that, despite current hybridization, reproductive isolation which maintains evolutionary boundaries between the two species, exists. Possible causes of hybridization phenomena are attempted, as well as their evolutionary and ecological implications, also considering a sea warming scenario in European waters.
Keywords: Anisakis; DNA microsatellites; SNPs; hybridization; mitochondrial introgression; mtDNA cox2.
Conflict of interest statement
The authors declare there are no conflicts of interest.
Figures




References
-
- Abattouy N, Valero A, Lozano J, Barón SD, Romero C and Martín-Sánchez J (2016) Population genetic analysis of Anisakis simplex s.l. and Anisakis pegreffii (Nematoda, Anisakidae) from parapatric areas and their contact zone. Parasite Epidemiology and Control 1, 169–176. doi: 10.1016/j.parepi.2016.02.003 - DOI - PMC - PubMed
-
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez‐Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Väinölä R, Wolf JBW and Zinner D (2013) Hybridization and speciation. Journal of Evolutionary Biology 26(2), 229–246. doi: 10.1111/j.1420-9101.2012.02599.x - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

