SRSF2 overexpression induces transcription-/replication-dependent DNA double-strand breaks and interferes with DNA repair pathways to promote lung tumor progression
- PMID: 40181846
- PMCID: PMC11963763
- DOI: 10.1093/narcan/zcaf011
SRSF2 overexpression induces transcription-/replication-dependent DNA double-strand breaks and interferes with DNA repair pathways to promote lung tumor progression
Abstract
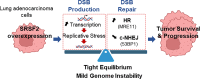
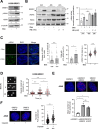
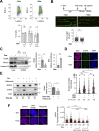
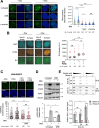
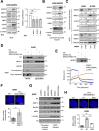
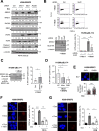
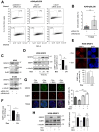
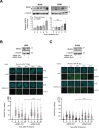
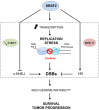
SRSF2 (serine/arginine-rich splicing factor 2) is a critical regulator of pre-messenger RNA splicing, which also plays noncanonical functions in transcription initiation and elongation. Although elevated levels of SRSF2 are associated with advanced stages of lung adenocarcinoma (LUAD), the mechanisms connecting SRSF2 to lung tumor progression remain unknown. We show that SRSF2 overexpression increases global transcription and replicative stress in LUAD cells, which correlates with the production of DNA damage, notably double-strand breaks (DSBs), likely resulting from conflicts between transcription and replication. Moreover, SRSF2 regulates DNA repair pathways by promoting homologous recombination and inhibiting nonhomologous end joining. Mechanistically, SRSF2 interacts with and enhances MRE11 (meiotic recombination 11) recruitment to chromatin, while downregulating 53BP1 messenger RNA and protein levels. Both events are likely contributing to SRSF2-mediated DNA repair process rerouting. Lastly, we show that SRSF2 and MRE11 expression is commonly elevated in LUAD and predicts poor outcome of patients. Altogether, our results identify a mechanism by which SRSF2 overexpression promotes lung cancer progression through a fine control of both DSB production and repair. Finally, we show that SRSF2 knockdown impairs late repair of ionizing radiation-induced DSBs, suggesting a more global function of SRSF2 in DSB repair by homologous recombination.
© The Author(s) 2025. Published by Oxford University Press on behalf of NAR Cancer.
Conflict of interest statement
None declared.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

