CDC42 Regulatory Patterns Related To Inflammatory Bowel Disease and Hyperglycemia
- PMID: 40183002
- PMCID: PMC11967731
CDC42 Regulatory Patterns Related To Inflammatory Bowel Disease and Hyperglycemia
Abstract
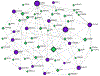
As a member of the rat sarcoma virus homolog (Rho) guanosine triphosphatases (GTPases) family, Cdc42 represents a "switch" molecule, by changing from inactive (GDP-associated) to active form (GTP-associated) and vice versa. Cdc42 is activated by the guanine nucleotide exchange factors (GEFs), in contrast to GTPase-activating proteins (GAPs) which are responsable for formation of GDP-binding, inactive form of Cdc42. Some of the fundamental cellular functions are regulated by Cdc42 such as cytosceleton dynamics, cell cycling, transcription and cellular trafficking. In the gastrointestinal system, Cdc42 participates in maintenance of the functional epithelial barrier by controling intestinal epithelial cell polarity and interconnections. In addition, Cdc42 expression in pancreatic β-cells is of great importance for glucose-stimulated insulin secretion. From the pathophysiological point of view, literature data provide some evidence for Cdc42 sigaling in inflammatory bowel disease, as well as in hyperglycemic conditions related to diabetes mellitus. However, whether and by which mechanism Cdc42 contributes to the IBD patophysiology in hyperglycemic conditions is still not fully understood. Therefore, we performed bioinformatics analysis to predict transcriptional factor-gene interactions related to Cdc42 signaling in inflammatory bowel disease in hyperglycemic conditions. In silico analysis predicts various interactions between input genes and output transcriptional factors, and therefore reveals the molecules with the highest predicted effect on particular genes. Based on the predictive interactions with the intracellular molecules, carefully designed in vitro or in vivo studies are required to get better insight in the pathways of interest. Better understanding of Cdc42 molecular pathway in inflammatory bowel disease and hyperglycemia will help identifying potential targets for therapeutical modifications in clinical setting resulting in better control of the disease progression.
Keywords: Cdc42; Diabetes; GTPase; GTPase-activating protein; Guanine nucleotide exchange factor; Guanosine triphosphatases; Hyperglycemia; Inflammatory bowel disease; Network analysis; Small GTPase; Transcription factors.
Conflict of interest statement
Competing interests: All authors have read the manuscript and declare no conflict of interest. No writing assistance was utilized in the production of this manuscript.
Figures



References
-
- Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS letters 582 (2008): 2093–2101. - PubMed
-
- Erickson JW, Zhang C-j, Kahn RA, et al. Mammalian Cdc42 is a brefeldin A-sensitive component of the Golgi apparatus. J Biol Chem 271 (1996): 26850–26854. - PubMed
-
- Wu WJ, Erickson JW, Lin R, et al. The γ-subunit of the coatomer complex binds Cdc42 to mediate transformation. Nature 405 (2000): 800–804. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
