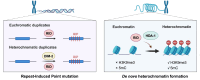
RID is required for both repeat-induced point mutation and nucleation of a novel transitional heterochromatic state for euchromatic repeats
- PMID: 40183634
- PMCID: PMC11969663
- DOI: 10.1093/nar/gkaf263
RID is required for both repeat-induced point mutation and nucleation of a novel transitional heterochromatic state for euchromatic repeats
Abstract
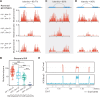
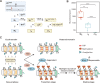
To maintain genome integrity, repeat sequences are subject to heterochromatin inactivation and, in Neurospora, repeat-induced point mutation (RIP). The initiating factors behind both are poorly understood. We resolve the paradoxical observation that newly introduced Repeat-Linker-Repeat (R-L-R) constructs require RID alone for RIP, while genomic repeats are RIPed in the absence of RID, showing that eu- and hetero- chromatic repeats are handled differently, the latter additionally requiring DIM-2. The differences between mechanisms associated with older and newer duplicates caution against extrapolation from mechanisms inferred from model experimental systems. Additionally, while chromatin status affects RIP, we also show that RID, when tethered with LexA, acts as a nucleation center for the transition from euchromatin to heterochromatin in an HDA-1 dependent fashion. Constitutive heterochromatin by contrast is largely HDA1 independent and depends on HDA-1 paralogs. RID is thus a dual function initiator of both RIP and the transition to heterochromatin.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
All authors beyond LDH declare no competing interests. L.D.H. is on the Scientific Advisory Board and has share options in ExpressionEdits.
Figures



Similar articles
-
DNA sequence homology induces cytosine-to-thymine mutation by a heterochromatin-related pathway in Neurospora.Nat Genet. 2017 Jun;49(6):887-894. doi: 10.1038/ng.3857. Epub 2017 May 1. Nat Genet. 2017. PMID: 28459455 Free PMC article.
-
Induction of H3K9me3 and DNA methylation by tethered heterochromatin factors in Neurospora crassa.Proc Natl Acad Sci U S A. 2017 Nov 7;114(45):E9598-E9607. doi: 10.1073/pnas.1715049114. Epub 2017 Oct 23. Proc Natl Acad Sci U S A. 2017. PMID: 29078403 Free PMC article.
-
Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa.Genome Res. 2009 Mar;19(3):427-37. doi: 10.1101/gr.086231.108. Epub 2008 Dec 17. Genome Res. 2009. PMID: 19092133 Free PMC article.
-
RIP: the evolutionary cost of genome defense.Trends Genet. 2004 Sep;20(9):417-23. doi: 10.1016/j.tig.2004.07.007. Trends Genet. 2004. PMID: 15313550 Review.
-
Genetic and epigenetic inactivation of repetitive sequences in Neurospora crassa: RIP, DNA methylation, and quelling.Curr Top Microbiol Immunol. 1995;197:165-77. doi: 10.1007/978-3-642-79145-1_11. Curr Top Microbiol Immunol. 1995. PMID: 7493491 Review. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

