Newly designed amplicons-based method for near-full-length genome (NFLG) sequencing of HIV-1 group M recombinant forms
- PMID: 40183829
- PMCID: PMC11971223
- DOI: 10.1007/s11033-025-10470-x
Newly designed amplicons-based method for near-full-length genome (NFLG) sequencing of HIV-1 group M recombinant forms
Abstract
Background: Over the years the spread of HIV-1 across the globe resulted in the creation of multiple subtypes and new recombinant forms (RFs). While the pol gene region of the HIV-1 genome is used for resistance mutations analysis and initial detection of RFs, whole genome sequencing analysis is required to determine recombination events across the viral genome. Here, we present a newly designed robust near-full length genome (NFLG) sequencing approach for the sequencing of HIV-1 genomes, out of clinical whole blood samples. This method has been successfully tested for various HIV-1 subtypes and RFs.
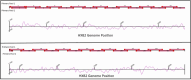
Methods and results: The method is based on an in-house developed set of 32 pan-genotypic primer pairs, divided into two pools, each containing 16 primer pairs covering the entire HIV-1 genome. Two parallel multiplex PCR reactions were used to generate 32 overlapping DNA fragments spanning the HIV-1 genome. Nextera XT protocol was used to obtain barcoded DNA libraries, which were sequenced with the Illumina Miseq platform using a V3 kit. A consensus sequence was determined for each sample and was used to define recombination events across the genome. For this aim, a combined analysis of several computational tools including HIV BLAST, phylogenetic analysis, RIP, SimPlot + + and jpHMM were employed. Overall, plasma samples from 33 patients suspected to carry RFs and 2 different, known pure subtypes controls, were included in this study. Genome coverage varied between RFs, while the gag and pol genes were nearly fully covered, the highly variable env gene region was not. Yet, these NFLG analyses enabled the identification of recombination events genome wide.
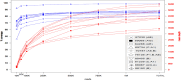
Conclusions: In summary, we describe a methodology for HIV-1 NFLG sequencing, which is based on partially overlapping, multiple PCR fragments, spanning the HIV-1 genome. Additionally, this newly refined method was compared to HIV-1 NFLG results of PCR-free metagenomic sequencing and proved to obtain greater coverage of the HXB2 reference genome. Yet, further testing and validation on a larger cohort is required. Still, this method enables sequencing of 20 different patient samples in a single MiSeq sequencing run and was used for the characterization of different HIV-1 RFs and pure subtypes circulating in Israel.
Keywords: Circulating recombinant forms (CRFs); HIV; HIV near-full length genome (NFLG) sequencing; HIV-1; Illumina miseq; Metagenomics; Next generation sequencing (NGS).
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no conflict of interest. Ethical approval: The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of the Sheba Medical Center (approval number 5803–18-SMC). Consent to participate: The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of the Sheba Medical Center (approval number 5803–18-SMC).
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

