A disease-specific convergence of host and Epstein-Barr virus genetics in multiple sclerosis
- PMID: 40184175
- PMCID: PMC12002260
- DOI: 10.1073/pnas.2418783122
A disease-specific convergence of host and Epstein-Barr virus genetics in multiple sclerosis
Abstract
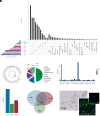
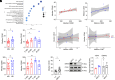
Recent sero-epidemiological studies have strengthened the hypothesis that Epstein-Barr virus (EBV) may be a causal factor in multiple sclerosis (MS). Given the complexity of the EBV-host interaction, various mechanisms may be responsible for the disease pathogenesis. Furthermore, it remains unclear whether this is a disease-specific process. Here, we showed that genes encoding EBV interactors are enriched in loci associated with MS but not with other diseases and in prioritized therapeutic targets. Analyses of MS blood and brain transcriptomes confirmed a dysregulation of MS-associated EBV interactors affecting the CD40 pathway. Such interactors were strongly enriched in binding sites for the EBV nuclear antigen 2 (EBNA2) viral transcriptional regulator, often in colocalization with CCCTC binding factor (CTCF) and RNA Polymerase II Subunit A (POLR2A). EBNA2 was expressed in the MS brain. The 1.2 EBNA2 allele downregulated the expression of the CD40 MS-associated gene analogously to the CD40 MS-risk variant. Finally, we showed that the 1.2 EBNA2 allele associates with the risk of MS. This study delineates how host and viral genetic variability converge in MS-specific pathogenetic mechanisms.
Keywords: Epstein–Barr virus; autoimmunity; multiple sclerosis.
Conflict of interest statement
Competing interests statement:M.S. and G.R. hold a patent on EBNA2 alleles in multiple sclerosis (IT1417523 EP2981625).
Figures


References
-
- Jacobs B. M., et al. , Towards a global view of multiple sclerosis genetics. Nat. Rev. Neurol. 18, 613–623 (2022). - PubMed
-
- Bjornevik K., et al. , Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science 375, 296–301 (2022). - PubMed
-
- Aloisi F., Giovannoni G., Salvetti M., Epstein-Barr virus as a cause of multiple sclerosis: Opportunities for prevention and therapy. Lancet Neurol. 22, 338–349 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

