VCF2Dis: an ultra-fast and efficient tool to calculate pairwise genetic distance and construct population phylogeny from VCF files
- PMID: 40184433
- PMCID: PMC11970368
- DOI: 10.1093/gigascience/giaf032
VCF2Dis: an ultra-fast and efficient tool to calculate pairwise genetic distance and construct population phylogeny from VCF files
Abstract
Background: Genetic distance metrics are crucial for understanding the evolutionary relationships and population structure of organisms. Progress in next-generation sequencing technology has given rise of genotyping data of thousands of individuals. The standard Variant Call Format (VCF) is widely used to store genomic variation information, but calculating genetic distance and constructing population phylogeny directly from large VCF files can be challenging. Moreover, the existing tools that implement such functions remain limited and have low performance in processing large-scale genotype data, especially in the area of memory efficiency.
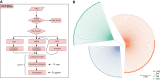
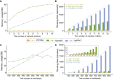
Findings: To address these challenges, we introduce VCF2Dis, an ultra-fast and efficient tool that calculates pairwise genetic distance directly from large VCF files and then constructs distance-based population phylogeny using the ape package. Benchmarking results demonstrate the tool's efficiency, with rapid processing times, minimal memory usage (e.g., 0.37 GB for the complete analysis of 2,504 samples with 81.2 million variants), and high accuracy, even when handling datasets with millions of variants from thousands of individuals. Its straightforward command-line interface, compatibility with downstream phylogenetic analysis tools (e.g., MEGA, Phylip, and FastTree), and support for multithreading make it a valuable tool for researchers studying population relationships. These advantages meaning VCF2Dis has already been widely utilized in many published genomic studies.
Conclusion: We present VCF2Dis, a straightforward and efficient tool for calculating genetic distance and constructing population phylogeny directly from large-scale genotype data. VCF2Dis has been widely applied, facilitating the exploration of population relationship in extensive genome sequencing studies.
Keywords: VCF; VCF2Dis; p-distance; population phylogeny.
© The Author(s) 2025. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare no potential competing interests.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

