Mg2+ influx mediated by TRPM7 triggers the initiation of muscle stem cell activation
- PMID: 40184450
- PMCID: PMC11970462
- DOI: 10.1126/sciadv.adu0601
Mg2+ influx mediated by TRPM7 triggers the initiation of muscle stem cell activation
Abstract
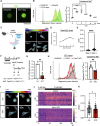
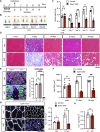
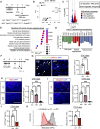
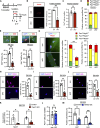
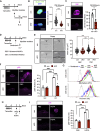
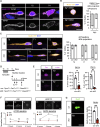
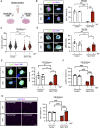
Muscle satellite cells (MuSCs) respond immediately to environmental cues upon skeletal muscle injuries. Despite decades of research into muscle regeneration, the specific molecular factors that trigger the transition of MuSCs from a quiescent to an active state remain largely unidentified. Here, we identify transient receptor potential melastatin 7 (TRPM7), an Mg2+-permeable ion channel, as a critical regulator of MuSC activation. Trpm7 deletion in MuSCs reduced Mg2+ influx, impairing myofiber regeneration and leading to decreased MuSC numbers and cell cycle arrest during regeneration. These changes were linked to disrupted mTOR signaling, which drives the transition of MuSCs from G0 to GAlert phase. In addition, Trpm7-deficient MuSCs exhibited impaired early responses, including quiescent projection retraction and AP-1 induction. Mg2+ supplementation rescued these defects, restoring normal MuSC activation. Our findings reveal a previously unrecognized mechanism where Mg2+ permeation through TRPM7 is essential for MuSC activation and efficient skeletal muscle regeneration, highlighting TRPM7 as a critical regulator of muscle repair.
Figures
References
-
- Reinhart R. A., Magnesium metabolism. A review with special reference to the relationship between intracellular content and serum levels. Arch. Intern. Med. 148, 2415–2420 (1988). - PubMed
-
- de Baaij J. H. F., Hoenderop J. G. J., Bindels R. J. M., Magnesium in man: Implications for health and disease. Physiol. Rev. 95, 1–46 (2015). - PubMed
-
- Schaffers O. J. M., Hoenderop J. G. J., Bindels R. J. M., de Baaij J. H. F., The rise and fall of novel renal magnesium transporters. Am. J. Physiol. Renal. Physiol. 314, F1027–F1033 (2018). - PubMed
-
- Runnels L. W., Yue L., Clapham D. E., TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science 291, 1043–1047 (2001). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

