Atf3 controls transitioning in female mitochondrial cardiomyopathy as identified by spatial and single-cell transcriptomics
- PMID: 40184463
- PMCID: PMC11970478
- DOI: 10.1126/sciadv.adq1575
Atf3 controls transitioning in female mitochondrial cardiomyopathy as identified by spatial and single-cell transcriptomics
Abstract
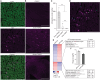
Oxidative phosphorylation defects result in now intractable mitochondrial diseases (MD) with cardiac involvement markedly affecting prognosis. The mechanisms underlying the transition from compensation to dysfunction in response to metabolic deficiency remain unclear. Here, we used spatially resolved transcriptomics and single-nucleus RNA sequencing (snRNA-seq) on the heart of a patient with mitochondrial cardiomyopathy (MCM), combined with an MCM mouse model with cardiac-specific Ndufs6 knockdown (FS6KD). Cardiomyocytes demonstrated the most heterogeneous expression landscape among cell types caused by metabolic perturbation, and pseudotime trajectory analysis revealed dynamic cellular states transitioning from compensation to severe compromise. This progression coincided with the transient up-regulation of a transcription factor, ATF3. Genetic ablation of Atf3 in FS6KD corroborated its pivotal role, effectively delaying cardiomyopathy progression in a female-specific manner. Our findings highlight a fate-determining role of ATF3 in female MCM progression and that the latest transcriptomic analysis will help decipher the mechanisms underlying MD progression.
Figures





References
-
- Gorman G. S., Chinnery P. F., DiMauro S., Hirano M., Koga Y., McFarland R., Suomalainen A., Thorburn D. R., Zeviani M., Turnbull D. M., Mitochondrial diseases. Nat. Rev. Dis. Prim. 2, 16080 (2016). - PubMed
-
- Gorman G. S., Schaefer A. M., Ng Y., Gomez N., Blakely E. L., Alston C. L., Feeney C., Horvath R., Yu-Wai-Man P., Chinnery P. F., Taylor R. W., Turnbull D. M., McFarland R., Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann. Neurol. 77, 753–759 (2015). - PMC - PubMed
-
- Suomalainen A., Battersby B. J., Mitochondrial diseases: The contribution of organelle stress responses to pathology. Nat. Rev. Mol. Cell Biol. 19, 77–92 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

