Essentiality of the virulence plasmid-encoded factors in disease pathogenesis of the major lineage of hypervirulent Klebsiella pneumoniae varies in different infection niches
- PMID: 40184910
- PMCID: PMC12002934
- DOI: 10.1016/j.ebiom.2025.105683
Essentiality of the virulence plasmid-encoded factors in disease pathogenesis of the major lineage of hypervirulent Klebsiella pneumoniae varies in different infection niches
Abstract
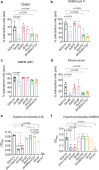
Background: Hypervirulent Klebsiella pneumoniae (HvKp) can metastasise to extra-intestinal sites to cause disseminated disease such as pyogenic liver abscesses. HvKp harbours a large virulence plasmid (KpVP) that contributes to pathogenicity. We previously identified a crucial gene region that confers virulence in SGH10 (ST23, K1 capsule), spanning genes encoding the siderophores aerobactin and salmochelin, as well as the regulator of mucoidy phenotype A (iuc-rmp-iro).
Methods: SGH10 isogenic mutants of aerobactin, rmpA, and salmochelin were generated and tested in vitro for their siderophore production, hypermucoviscosity and growth. We investigated the essentiality of these factors in different murine infection or colonisation models.
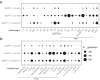
Findings: In a lung pneumonia model, capsule modulation by rmpA was the primary driver of high bacterial burden in the lung. In a systemic infection setting, rmpA was still the primary driver, followed by a significant contribution by salmochelin, that conferred virulence. However, the role of aerobactin was more significant in hvKp persistence in the gut. We further examined a large collection of Kp genomes and observed that the iro loci is often co-inherited with iuc in KpVP-1, suggesting the evolutionary importance of expressing both siderophores in these lineages.
Interpretation: HvKp typically colonises the intestinal niche, however, the acquisition of the KpVP plasmid has enabled it to thrive outside the gut and cause metastatic infections. While the iuc-rmp-iro region is pivotal in bestowing virulence, the encoded factors contribute differently to the success of the pathogen in various infection sites, where the microenvironment, nutrient availability and immune response can vary. Thus, our study demonstrates that possessing the iuc-rmp-iro gene region can be an evolutionary advantage by allowing for flexibility in modulating siderophore and capsule expression in order for K. pneumoniae to thrive in distinct host niches.
Funding: This work is funded by the National Research FoundationMOH-000925-00 to YH Gan and OFYIRG22jul-0042 by the National Medical Research Council (NMRC) to THT.
Keywords: Hypervirulent; Klebsiella pneumoniae; Liver abscess; Plasmid; Siderophore.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflicts of interest.
Figures



References
-
- Murray C.J.L. The global burden of disease study at 30 years. Nat Med. 2022;28(10):2019–2026. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

