Gut microbiota diversity among humans, elephants, livestock and wild herbivores in Chitwan National Park bears implications for conservation medicine
- PMID: 40185849
- PMCID: PMC11971256
- DOI: 10.1038/s41598-025-89402-5
Gut microbiota diversity among humans, elephants, livestock and wild herbivores in Chitwan National Park bears implications for conservation medicine
Abstract
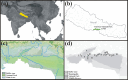
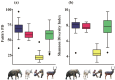
Gut microbiome influences host health and well-being. Co-occurring hosts may exchange disease-causing bacteria belonging to these microbial communities. Therefore, monitoring gut microbiota composition in wildlife and humans is paramount to prevent zoonotic diseases, thus protecting and strengthening public health. We characterized diversity and abundance of the gut microbiome bacterial component across mahouts (captive elephant trainers and handlers), their pachyderms, livestock and wild herbivores in and around Chitwan National Park (Nepal). Firmicutes and Bacteroidota were invariably the dominant phyla. In humans, the relative abundance of Firmicutes was higher, the alpha diversity lower and beta diversity different compared to other host categories. Livestock and wild herbivores displayed similar alpha and beta diversity due to the presence of Proteobacteria, Actinobacteriota and Verrucomicrobiota. Elephants had a higher alpha diversity, and a significant beta diversity compared to other mammals. Our results suggest that taxonomic affiliation and diet niche are the main drivers of gut microbiota composition. Nevertheless, Mycobacterium and other potentially pathogenic bacteria genera were detected in elephants and livestock other than wild herbivores. These findings shed light on microbiota sharing and interlinking in each environment, thereby highlighting the importance of conservation medicine to better our understanding of health in co-occurring host species.
Keywords: Bacterial function; Human–wildlife–livestock–environment interface; Microbiome; Nepal; One Health; Zoonosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics statement: The ethical approval of the study was provided by the Nepal Ethical Review Board of Nepal Health Research Council (IRC number 312/2018). All individuals gave written informed consent before participation during the study. The experimental protocol used in the present study was approved by the Bioethics Committee of BIOPOLIS/CIBIO-InBIO (University of Porto). Studies were conducted in accordance with the principles of the Declaration of Helsinki.
Figures


Similar articles
-
A comparison of wild boar and domestic pig microbiota does not reveal a loss of microbial species but an increase in alpha diversity and opportunistic genera in domestic pigs.Microbiol Spectr. 2024 Oct 3;12(10):e0084324. doi: 10.1128/spectrum.00843-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162552 Free PMC article.
-
Characteristics of gut microbiota profiles in Asian elephants (Elephas maximus) with gastrointestinal disorders.Sci Rep. 2025 Jan 8;15(1):1327. doi: 10.1038/s41598-025-85495-0. Sci Rep. 2025. PMID: 39779898 Free PMC article.
-
Distinct fecal microbiome between wild and habitat-housed captive polar bears (Ursus maritimus): Impacts of captivity and dietary shifts.PLoS One. 2024 Nov 20;19(11):e0311518. doi: 10.1371/journal.pone.0311518. eCollection 2024. PLoS One. 2024. PMID: 39565828 Free PMC article.
-
Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates.Sci Total Environ. 2022 Sep 10;838(Pt 2):156178. doi: 10.1016/j.scitotenv.2022.156178. Epub 2022 May 23. Sci Total Environ. 2022. PMID: 35618126 Review.
-
Using the microbiota to study connectivity at human-animal interfaces.Trends Microbiol. 2025 Jun 9:S0966-842X(25)00153-2. doi: 10.1016/j.tim.2025.05.006. Online ahead of print. Trends Microbiol. 2025. PMID: 40494694 Review.
References
-
- D’Argenio, V. & Salvatore, F. The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta451, 97–102. 10.1016/J.CCA.2015.01.003 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

