The tandem duplicator phenotype may be a novel targetable subgroup in pancreatic cancer
- PMID: 40185871
- PMCID: PMC11971333
- DOI: 10.1038/s41698-025-00888-8
The tandem duplicator phenotype may be a novel targetable subgroup in pancreatic cancer
Abstract
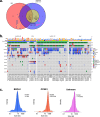
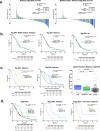
Tandem duplicator phenotype (TDP) consists of distinct genomic rearrangements where tandem duplications are randomly distributed. In this study, we characterized the prevalence and outcomes of TDP in a large series of prospectively sequenced tumors from patients with pancreatic ductal adenocarcinomas (PDAC). Whole-genome sequencing (WGS) was performed in 530 PDAC cases from the PanCuRx Initiative, COMPASS and PanGen/POG trials in Canada. Of 530 cases, 52 were identified as TDP (9.8%; 13 resected, 39 advanced). Etiological subgroups of TDP included BRCA1 (n = 9), CCNE1 (n = 4), and unknown (n = 39). Presence of TDP was not prognostic in resected specimens (p = 0.77) compared with non-HRD and non-TDP cases, described as typicals. In advanced cases, when stratified for only classical subtype cases, platinum therapy was correlated with longer response in non-BRCA1 TDP vs. typicals (p = 0.0036). There was no difference in overall survival between TDP and typicals (p = 0.5).TDP represents a potential novel targetable subgroup for chemotherapy selection in PDAC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors declare no competing interests.
Figures


References
-
- Siegel, R.L., Miller, K.D. & Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin.66, 7–30 (2016). - PubMed
-
- Pishvaian M. J., et al. Outcomes in patients with pancreatic Adenocarcinoma with genetic mutations in DNA damage response pathways: results from the know your tumor program. JCO Precis. Oncol. 1–10 (2019). - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

