Hepatocyte TM4SF5-mediated cytosolic NCOA3 stabilization and macropinocytosis support albumin uptake and bioenergetics for hepatocellular carcinoma progression
- PMID: 40186033
- PMCID: PMC12046047
- DOI: 10.1038/s12276-025-01438-9
Hepatocyte TM4SF5-mediated cytosolic NCOA3 stabilization and macropinocytosis support albumin uptake and bioenergetics for hepatocellular carcinoma progression
Abstract
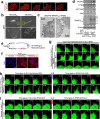
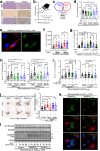
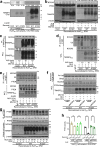
Transmembrane 4 L six family member 5 (TM4SF5) is involved in hepatocellular carcinoma (HCC) development and progression. Although TM4SF5 also promotes migration and invasion, it remains unclear how the metabolic context affects metastatic potential. Here we explored how TM4SF5 affects albumin uptake for HCC progression using TM4SF5 knockout or reintroduced hepatocyte and animal systems. Serum-deprived hepatocytes formed filopodia-like processes depending on TM4SF5 expression, which was altered by albumin replenishment for membranous PIP3-dependent macropinocytosis. Macropinocytosis required nuclear receptor coactivator 3 (NCOA3) stabilized in the cytosol and PTEN inactivation via binding to TM4SF5WT. TM4SF5-mediated albumin uptake led to ATP-linked respiration and cellular migration. Tumor tissues from liver-orthotopically xenografted mice fed a high protein diet or human liver cancer tissues showed TM4SF5-dependent macropinocytosis and NCOA3-correlated metastatic features, unlike mice fed a normal chow diet or human nontumor regions. These observations indicate that serum albumin availability to TM4SF5-positive HCC could support multifocality and intrahepatic metastasis, which may provide insights into clinical observations of multiple small tumor nodules surrounded by areas with high serum albumin levels.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors have no conflicts of interest to declare.
Figures





References
-
- Young, L. B. et al. Survival analysis using albumin–bilirubin (ALBI) grade for patients treated with drug-eluting embolic transarterial chemoembolization for hepatocellular carcinoma. J. Vasc. Interv. Radiol.33, 510–517.e511 (2022). - PubMed
-
- Vauthey, J. N. et al. Simplified staging for hepatocellular carcinoma. J. Clin. Oncol.20, 1527–1536 (2002). - PubMed
-
- Spinella, R., Sawhney, R. & Jalan, R. Albumin in chronic liver disease: structure, functions and therapeutic implications. Hepatol. Int10, 124–132 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

