The Chlamydia effector Dre1 binds dynactin to reposition host organelles during infection
- PMID: 40186871
- PMCID: PMC12139634
- DOI: 10.1016/j.celrep.2025.115509
The Chlamydia effector Dre1 binds dynactin to reposition host organelles during infection
Abstract
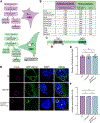
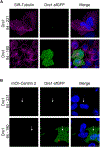
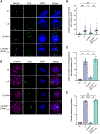
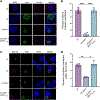
The obligate intracellular pathogen Chlamydia trachomatis replicates in a specialized membrane-bound compartment where it repositions host organelles during infection to acquire nutrients and evade host surveillance. We describe a bacterial effector, Dre1, that binds specifically to dynactin associated with host microtubule organizing centers without globally impeding dynactin function. Dre1 is required to reposition the centrosome, mitotic spindle, Golgi apparatus, and primary cilia around the inclusion and contributes to pathogen fitness in cell-based and mouse models of infection. We utilized Dre1 to affinity purify the megadalton dynactin protein complex and determined the first cryoelectron microscopy (cryo-EM) structure of human dynactin. Our results suggest that Dre1 binds to the pointed end of dynactin and uncovers the first bacterial effector that modulates dynactin function. Our work highlights how a pathogen employs a single effector to evoke targeted, large-scale changes in host cell organization that facilitate pathogen growth without inhibiting host viability.
Keywords: CP: Cell biology; CP: Microbiology; Chlamydia; Golgi apparatus; bacterial pathogenesis; centrosome; dynactin; host-pathogen interactions; microtubule organizing center; primary cilia.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The Krogan Laboratory has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. N.J.K. has a financially compensated consulting agreement with Maze Therapeutics. He is the President of and is on the Board of Directors of Rezo Therapeutics, and he is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, GEn1E Lifesciences, and Interline Therapeutics. R.H.V. is co-founder at Bloom Sciences (San Diego, CA, USA), a microbiome-based therapeutics company. The company was not involved in funding or interpreting the work presented.
Figures


References
-
- Bennett JE, Dolin R, Blaser MJ, and Mandell D (2020). Bennett’s Principles and Practice of Infectious Diseases Ninth Edition (Elsevier; ).
-
- World Health Organization (2016). WHO guidelines for the treatment of chlamydia trachomatis (World Health Organization; ). https://iris.who.int/handle/10665/246165. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

