ASCL1 protein domains with distinct functions in neuronal differentiation and subtype specification
- PMID: 40187474
- PMCID: PMC12535295
- DOI: 10.1016/j.ydbio.2025.04.001
ASCL1 protein domains with distinct functions in neuronal differentiation and subtype specification
Abstract
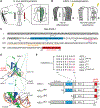
ASCL1 is a neural basic helix-loop-helix (bHLH) transcription factor that plays essential roles during neural development, including neural differentiation and neuronal subtype specification. bHLH factors are defined by their motifs, including a basic region interacting with DNA and an HLH domain involved in protein-protein interactions. We previously defined specific regions within the bHLH domain of ASCL1 as important for its specific functions directing neuronal differentiation in the chick neural tube. Here, we build upon these findings to show how specific mutations within the basic region block DNA binding but not heterodimer formation with E-protein partners TCF3 (E12/E47) and TCF12 (HEB) yet have differential abilities to show dominant negative phenotypes. Additionally, truncating domains outside the bHLH define a nuclear localization signal, a requirement for the C-terminal acidic residues, and the non-essentiality of the N-terminal glutamine/alanine repeats. This structure/function analysis identifies functional domains for ASCL1 activity.
Keywords: ASCL1; Neurogenesis; Neuronal specification; Spinal cord development; bHLH transcription factors.
Copyright © 2025 Elsevier Inc. All rights reserved.
Figures






References
-
- Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, Bodenstein SW, Evans DA, Hung CC, O’Neill M, Reiman D, Tunyasuvunakool K, Wu Z, Zemgulyte A, Arvaniti E, Beattie C, Bertolli O, Bridgland A, Cherepanov A, Congreve M, Cowen-Rivers AI, Cowie A, Figurnov M, Fuchs FB, Gladman H, Jain R, Khan YA, Low CMR, Perlin K, Potapenko A, Savy P, Singh S, Stecula A, Thillaisundaram A, Tong C, Yakneen S, Zhong ED, Zielinski M, Zidek A, Bapst V, Kohli P, Jaderberg M, Hassabis D, Jumper JM, 2024. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500. - PMC - PubMed
-
- Battiste J, Helms AW, Kim EJ, Savage TK, Lagace DC, Mandyam CD, Eisch AJ, Miyoshi G, Johnson JE, 2007. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development 134, 285–293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

