A differential requirement for ciliary transition zone proteins in human and mouse neural progenitor fate specification
- PMID: 40188187
- PMCID: PMC11972330
- DOI: 10.1038/s41467-025-58554-3
A differential requirement for ciliary transition zone proteins in human and mouse neural progenitor fate specification
Abstract
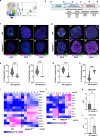
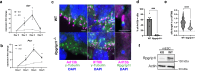
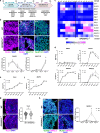
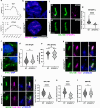
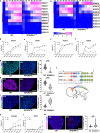
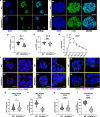
Studying ciliary genes in the context of the human central nervous system is crucial for understanding the underlying causes of neurodevelopmental ciliopathies. Here, we use pluripotent stem cell-derived spinal organoids to reveal distinct functions of the ciliopathy gene RPGRIP1L in humans and mice, and uncover an unexplored role for cilia in human axial patterning. Previous research has emphasized Rpgrip1l critical functions in mouse brain and spinal cord development through the regulation of SHH/GLI pathway. Here, we show that RPGRIP1L is not required for SHH activation or motoneuron lineage commitment in human spinal progenitors and that this feature is shared by another ciliopathy gene, TMEM67. Furthermore, human RPGRIP1L-mutant motoneurons adopt hindbrain and cervical identities instead of caudal brachial identity. Temporal transcriptome analysis reveals that this antero-posterior patterning defect originates in early axial progenitors and correlates with cilia loss. These findings provide important insights into the role of cilia in human neural development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
- WI 5451/1-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- ANR-17-CE16-0003-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-17-RHUS-0002/Agence Nationale de la Recherche (French National Research Agency)
- ANR-10-IAHU-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-19-CE16-0002/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources

