Cytogenetic signatures favoring metastatic organotropism in colorectal cancer
- PMID: 40188208
- PMCID: PMC11972295
- DOI: 10.1038/s41467-025-58413-1
Cytogenetic signatures favoring metastatic organotropism in colorectal cancer
Abstract
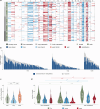
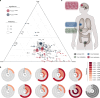
Colorectal carcinoma (CRC) exhibits metastatic organotropism, primarily targeting liver, lung, and rarely the brain. Here, we study chromosomal imbalances (CIs) in cohorts of primary CRCs and metastases. Brain metastases show the highest burden of CIs, including aneuploidies and focal CIs, with enrichment of +12p encoding KRAS. Compared to liver and lung metastases, brain metastases present with increased co-occurrence of KRAS mutation and amplification. CRCs with concurrent KRAS mutation and amplification display significant metabolic reprogramming with upregulation of glycolysis, alongside upregulation of cell cycle pathways, including copy number gains of MDM2 and CDK4. Evolutionary modeling suggests early acquisition of many organotropic CIs enriched in both liver and brain metastases, while brain-enriched CIs preferentially emerge later. Collectively, this study supports a model where cytogenetic events in CRCs favor site-specific metastatic colonization. These site-enriched CI patterns may serve as biomarkers for metastatic potential in precision oncology.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors are not aware of any conflict of interest related to this study.
Figures





References
-
- Global Burden of Disease Cancer Collaboration. et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study.JAMA Oncol5, 1749–1768 (2019). - PMC - PubMed
-
- Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science331, 1559–1564 (2011). - PubMed
-
- Elferink, M. A., de Jong, K. P., Klaase, J. M., Siemerink, E. J. & de Wilt, J. H. Metachronous metastases from colorectal cancer: a population-based study in North-East Netherlands. Int J. Colorectal Dis.30, 205–212 (2015). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

