A single amino acid in the Salmonella effector SarA/SteE triggers supraphysiological activation of STAT3 for anti-inflammatory gene expression
- PMID: 40188438
- PMCID: PMC12014907
- DOI: 10.1016/j.celrep.2025.115530
A single amino acid in the Salmonella effector SarA/SteE triggers supraphysiological activation of STAT3 for anti-inflammatory gene expression
Abstract
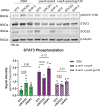
Salmonella causes ∼1 million cases of gastroenteritis annually in the United States. Critical to virulence are secreted effectors that reprogram host functions. We previously discovered the effector SarA facilitates phosphorylation of STAT3, inducing expression of the anti-inflammatory cytokine interleukin-10 (IL-10). This STAT3 activation requires a region of homology with the host cytokine receptor gp130. Here, we demonstrate that a single amino acid difference is critical for the anti-inflammatory bias of SarA-STAT3 signaling. An isoleucine at pY+1 of the YxxQ motif in SarA (which binds the STAT3 SH2 domain) causes increased STAT3 recruitment and phosphorylation, biasing toward anti-inflammatory targets. This isoleucine renders SarA a better substrate for tyrosine phosphorylation by GSK-3. GSK-3 is canonically a serine/threonine kinase that nonetheless undergoes tyrosine autophosphorylation at a motif with isoleucine at the pY+1 position. Our results provide a molecular basis for how a Salmonella effector achieves supraphysiological levels of STAT3 activation to control host genes.
Keywords: CP: Immunology; CP: Microbiology; GSK-3; IL-10; IL-6; IL6ST; JAK-STAT; SOCS3; STM2585; Y705; adaptation; gogC.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Update of
-
A single amino acid in the Salmonella effector SarA/SteE triggers supraphysiological activation of STAT3 for anti-inflammatory target gene expression.bioRxiv [Preprint]. 2024 Feb 14:2024.02.14.580367. doi: 10.1101/2024.02.14.580367. bioRxiv. 2024. Update in: Cell Rep. 2025 Apr 22;44(4):115530. doi: 10.1016/j.celrep.2025.115530. PMID: 38405869 Free PMC article. Updated. Preprint.
References
-
- Hoffmann S., Maculloch B., Batz M. Economic Burden of Major Foodborne Illnesses Acquired in the United States, EIB-140. U.S. Department of Agriculture, Economic Research Service; 2015.
-
- Galán J.E., Collmer A. Type III Secretion Machines: Bacterial Devices for Protein Delivery into Host Cells. Science. 1999;284:1322–1328. - PubMed
-
- Jaslow S.L., Gibbs K.D., Fricke W.F., Wang L., Pittman K.J., Mammel M.K., Thaden J.T., Fowler V.G., Jr., Hammer G.E., Elfenbein J.R., Ko D.C. Salmonella Activation of STAT3 Signaling by SarA Effector Promotes Intracellular Replication and Production of IL-10. Cell Rep. 2018;23:3525–3536. doi: 10.1016/j.celrep.2018.05.072. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

