Robust Assessment of Homologous Recombination Deficiency Genomic Instability by OncoScan Microarrays
- PMID: 40188947
- PMCID: PMC12163384
- DOI: 10.1016/j.jmoldx.2025.02.011
Robust Assessment of Homologous Recombination Deficiency Genomic Instability by OncoScan Microarrays
Abstract
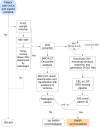
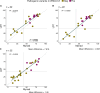
Genomic instability scars are markers for detecting homologous recombination deficiency (HRD) status in patients with ovarian cancer and predicting the response to poly (ADP-ribose) polymerase inhibitor treatment. Currently, only a few reliable and validated assays are available, with the Myriad myChoice CDx being the most commonly used commercial assay for genomic instability scar score determination. Given the need for a more straightforward, accessible, and reliable method for detecting genomic instability scars methods, in this work, we describe the feasibility of using the microarray OncoScan copy number variant assay and open-source software packages to quantify genomic instability scores, and the development of an open-access online platform for genomic instability score calculation. The laboratory-developed test accurately classified homologous recombination-proficient and recombination-deficient samples based on genomic instability scores derived from the OncoScan copy number variant assay. Internally evaluated genomic instability scores demonstrated a 92% overall agreement and a higher sample success rate compared with externally analyzed genomic instability scar scores. The availability of HRD determination has doubled the number of patients eligible for poly (ADP-ribose) polymerase therapy. The assay can be conveniently performed on individual samples, and the open-access online platform facilitates HRD determination without the need for specialized bioinformatics support.
Copyright © 2025 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure Statement None declared.
Figures


References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Varol U., Kucukzeybek Y., Alacacioglu A., Somali I., Altun Z., Aktas S., Oktay Tarhan M. BRCA genes: BRCA 1 and BRCA 2. J BUON. 2018;23:862–866. - PubMed
-
- Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.-A., Mooij T.M., Roos-Blom M.-J., et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402. - PubMed
-
- Miller R.E., Leary A., Scott C.L., Serra V., Lord C.J., Bowtell D., Chang D.K., Garsed D.W., Jonkers J., Ledermann J.A., Nik-Zainal S., Ray-Coquard I., Shah S.P., Matias-Guiu X., Swisher E.M., Yates L.R. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann Oncol. 2020;31:1606–1622. - PubMed
-
- Ray-Coquard I., Pautier P., Pignata S., Pérol D., González-Martín A., Berger R., Fujiwara K., Vergote I., Colombo N., Mäenpää J., Selle F., Sehouli J., Lorusso D., Guerra Alía E.M., Reinthaller A., Nagao S., Lefeuvre-Plesse C., Canzler U., Scambia G., Lortholary A., Marmé F., Combe P., De Gregorio N., Rodrigues M., Buderath P., Dubot C., Burges A., You B., Pujade-Lauraine E., Harter P. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381:2416–2428. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

