CpG site methylation regulates mouse Rec8 gene promoter activity
- PMID: 40189260
- PMCID: PMC12151637
- DOI: 10.1262/jrd.2024-077
CpG site methylation regulates mouse Rec8 gene promoter activity
Abstract
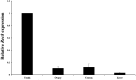
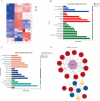
The Rec8 gene is specifically expressed in fetal and adult gonads. Although the importance of REC8 in gametogenesis is widely acknowledged, the mechanisms underlying its germ cell-specific expression remain unclear. In this study, we utilized the mouse Rec8 gene sequence to construct a 2577 bp sequence, which included intron 1 (180 bp), exon 1 (118 bp), and an upstream 2279 bp region. The dual-luciferase assay results showed significant differences in promoter activity between -650 bp and -385 bp and between -89 bp and -35 bp. This indicated that the core promoter region of the Rec8 gene may exist within these regions. Bisulfite sequencing PCR results showed that CpGs 10-19 were largely unmethylated in the testes but hypermethylated in other tissues. Interestingly, correlation analysis between CpG methylation status and Rec8 mRNA expression levels showed that methylation of CpGs 10 to 19 was negatively correlated with Rec8 mRNA expression levels (Pearson's r = -0.991, P = 0.009). Furthermore, RNA-Seq data and bioinformatic analyses suggested that the specific expression of Rec8 may be linked to the presence of TATA-like sequences within its core promoter region. Overall, these findings indicate that Rec8 expression is regulated by the low methylation of CpG sites and the presence of TATA-like sequences in its core promoter.
Keywords: Cohesin; DNA methylation; Gene expression; Promoter; Rec8.
Conflict of interest statement
The authors declare no competing interests
Figures



References
-
- Lee J, Iwai T, Yokota T, Yamashita M. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J Cell Sci 2003; 116: 2781–2790. - PubMed

