Aneuploidy confers a unique transcriptional and phenotypic profile to Candida albicans
- PMID: 40189588
- PMCID: PMC11973194
- DOI: 10.1038/s41467-025-58457-3
Aneuploidy confers a unique transcriptional and phenotypic profile to Candida albicans
Abstract
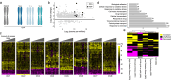
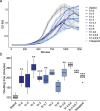
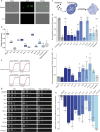
Inaccurate chromosome segregation can lead to the formation of aneuploid cells that harbor an imbalanced complement of chromosomes. Several fungal species are not only able to tolerate the detrimental effects of aneuploidy but can use it to adapt to environmental pressures. The fungal pathobiont Candida albicans frequently acquires supernumerary chromosomes that enable growth in the presence of antifungal drugs or in specific host niches, yet the transcriptional changes associated with aneuploidy are not globally defined. Here, a karyotypically diverse set of C. albicans strains revealed that expression generally correlated with gene copy number regardless of the strain karyotype. Unexpectedly, aneuploid strains shared a characteristic transcriptional profile that was distinct from a generalized environmental stress response previously defined in aneuploid yeast cells. This aneuploid transcriptional response led to altered growth and oxidative balances relative to euploid control strains. The increased expression of reactive oxygen species (ROS) mitigating enzymes in aneuploid cells reduced the levels of ROS but caused an acute sensitivity to both internal and external sources of oxidative stress. Taken together, our work demonstrates common transcriptional and phenotypic features of aneuploid C. albicans cells with consequences for infection of different host niches and susceptibility to environmental stimuli.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

